Methanol dehydrogenase (cytochrome c)
Methanol dehydrogenase (MEDH) is a periplasmic quinoprotein. It catalyses the oxidation of methanol and other small alcohols to the corresponding aldehyde with the release of two protons and two electrons. The enzyme uses the pyrroloquinolinequinone (PQQ) cofactor.
MEDH is an H2L2 heterotetramer, made of two heavy (H) chains and two light (L) chains. Each H subunit contains one molecule of the prosthetic group PQQ, which is non-covalently bound to the polypeptide chain, as well as one calcium ion which is catalytically essential. There is no interaction between the L chains which fold around the surface of the H chains.
Reference Protein and Structure
- Sequences
-
P38539
 (1.1.2.7)
(1.1.2.7)
P38540 (1.1.2.7)
(1.1.2.7)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Methylophilus methylotrophus (Bacteria)

- PDB
-
1g72
- CATALYTIC MECHANISM OF QUINOPROTEIN METHANOL DEHYDROGENASE: A THEORETICAL AND X-RAY CRYSTALLOGRAPHIC INVESTIGATION
(1.9 Å)



- Catalytic CATH Domains
-
2.140.10.10
 (see all for 1g72)
(see all for 1g72)
- Cofactors
- Calcium(2+) (1), Pyrroloquinoline quinone(3-) (1) Metal MACiE
Enzyme Mechanism
Introduction
In this computationally proposed mechanism, the reaction is initiated by the abstraction of a proton from the alcohol substrate by the general base Glu117. Simultaneously, a hydride is transferred to C'5 of PQQ, forming PQQH-. The aldehyde product then leaves the active site via an unknown pathway. The proton from Glu171 is transferred to Asp297, where it is positioned correctly to be abstracted by the alkoxide of PQQH-, forming PQQH. Quantum mechanical modelling then suggests Glu171 acts as a general base through a water molecule to initiate enolisation of PQQH to PQQH2. The presence of a calcium divalent cation within the active site polarises the PQQ carbonyl towards accepting a hydride.
The exact route that the electrons and protons take out of the active site to the heme of the bound cytochrome cL during the reduction of PQQ is unknown (due to lack of crystal structure with the cytochrome bound). It is assumed that the electrons will take the shortest route and may be the conserved disulfide ring, Asp105 and Asn52 and water molecules. The protons from the quinol are thought to be released to the periplasm through a hydrogen bonded network of proton donor and acceptor groups. This is thought to involve Asp303, Arg331. Glu177 and a channel of water molecules leading out of the active site [PMID:12686102].
Catalytic Residues Roles
| UniProt | PDB* (1g72) | ||
| Asp299 | Asp297(299)A | In the hydride mechanism, Asp297 is thought to act as a general acid to the PQQH- alkoxide anion, donating a proton relayed from Glu171. Modelling studies have ruled out Asp297 as the general base to initiate hydride transfer, although this has been suggested by structural data. | metal ligand, proton acceptor, proton donor |
| Asn257 | Asn255(257)A | Forms part of the calcium binding site. | metal ligand |
| Glu173 | Glu171(173)A | The residue is correctly positioned within the active site to act as a general base towards the alcohol substrate, initiating hydride transfer. It is thought to relay its attained proton to Asp297 which is positioned to act as a general acid to the alkoxide of PQQH-. The carboxylate anion of Glu171 is then thought to initiate enolisation of the PQQH prosthetic group to PQQH2 through a water molecule. | metal ligand, proton acceptor, proton donor |
Chemical Components
hydride transfer, proton transfer, proton relay, electron transfer, native state of cofactor regenerated, inferred reaction step, native state of enzyme regeneratedReferences
- Reddy SY et al. (2004), Protein Sci, 13, 1965-1978. Determination of enzyme mechanisms by molecular dynamics: Studies on quinoproteins, methanol dehydrogenase, and soluble glucose dehydrogenase. DOI:10.1110/ps.04673404. PMID:15273299.
- Rozeboom HJ et al. (2015), Protein Sci, 24, 2044-2054. Crystal structure of quinone-dependent alcohol dehydrogenase fromPseudogluconobacter saccharoketogenes. A versatile dehydrogenase oxidizing alcohols and carbohydrates. DOI:10.1002/pro.2818. PMID:26440996.
- Reddy SY et al. (2004), Proc Natl Acad Sci U S A, 101, 15887-15892. Mechanisms of ammonia activation and ammonium ion inhibition of quinoprotein methanol dehydrogenase: A computational approach. DOI:10.1073/pnas.0407209101. PMID:15520392.
- Anthony C et al. (2003), Biochim Biophys Acta, 1647, 18-23. The structure and mechanism of methanol dehydrogenase. DOI:10.1016/s1570-9639(03)00042-6. PMID:12686102.
- Xia ZX et al. (1999), Biochemistry, 38, 1214-1220. Detailed Active Site Configuration of a New Crystal Form of Methanol Dehydrogenase fromMethylophilusW3A1 at 1.9 Å Resolution†,‡. DOI:10.1021/bi9822574. PMID:9930981.
- Anthony C et al. (1994), Biochem J, 304, 665-674. The structure and function of methanol dehydrogenase and related quinoproteins containing pyrrolo-quinoline quinone. DOI:10.1042/bj3040665. PMID:7818466.

Step 1. Glu171 as the catalytic base, abstracting the alcohol hydroxyl proton and initiating hydride transfer to the C5 carbonyl of PQQ. The aldehyde product then leaves the active site via an unknown pathway.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asn255(257)A | metal ligand |
| Asp297(299)A | metal ligand |
| Glu171(173)A | metal ligand |
| Glu171(173)A | proton acceptor |
Chemical Components
hydride transfer, proton transferCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asn255(257)A | metal ligand |
| Asp297(299)A | metal ligand |
| Glu171(173)A | metal ligand |
| Asp297(299)A | proton acceptor |
| Glu171(173)A | proton donor |
Chemical Components
proton transfer
Step 3. Asp297, which is thought to be correctly positioned to act as a general acid towards the alkoxide anion of PQQH-, forming PQQH.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asn255(257)A | metal ligand |
| Asp297(299)A | metal ligand |
| Glu171(173)A | metal ligand |
| Asp297(299)A | proton donor |
Chemical Components
proton transfer
Step 4. Computer modelling studies, which support the alternative hydride mechanism suggest that Glu171 acts as a general base through a water molecule to initiate enolisation of PQQH to PQQH2.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asn255(257)A | metal ligand |
| Asp297(299)A | metal ligand |
| Glu171(173)A | metal ligand, proton acceptor |
Chemical Components
proton relay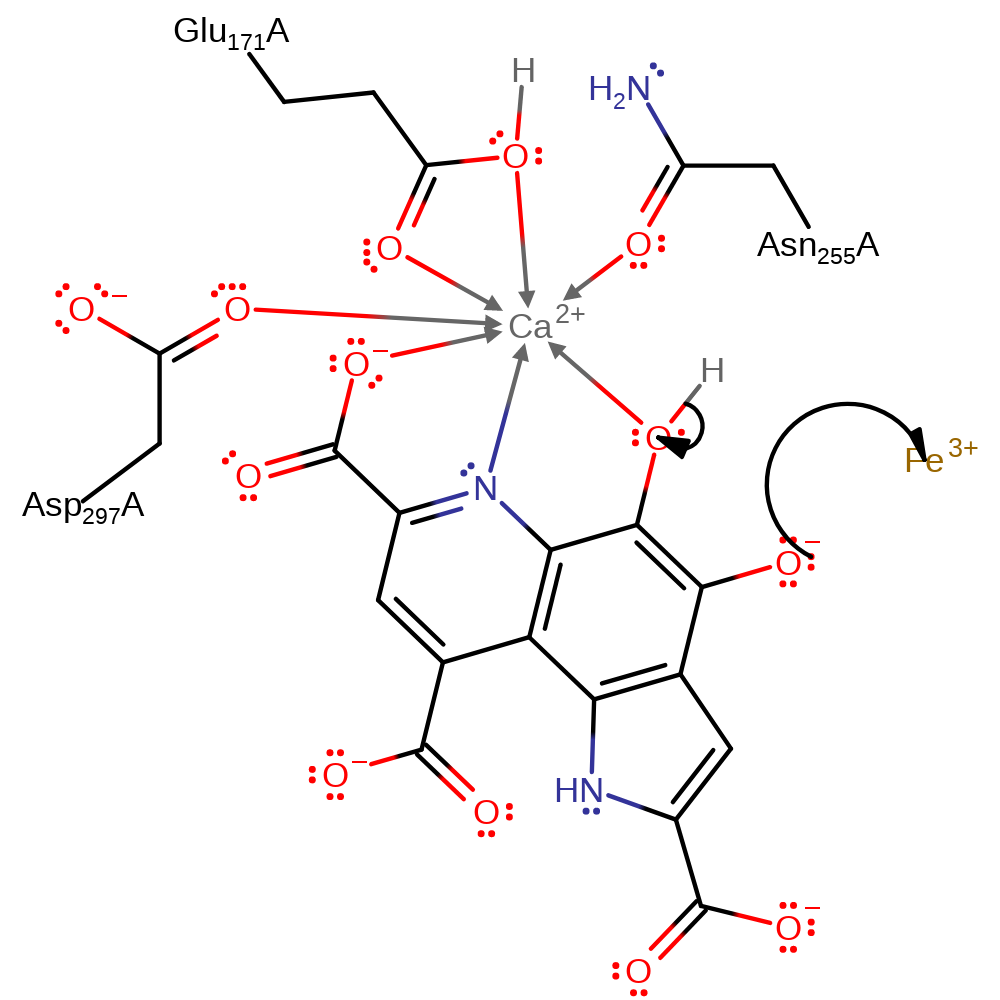
Step 5. In the first step of the reduction of PQQ, a single electron and single proton are lost from the cofactor.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asn255(257)A | metal ligand |
| Asp297(299)A | metal ligand |
| Glu171(173)A | metal ligand |
Chemical Components
electron transfer
Step 6. In the final step of the reduction of PQQ, a single electron is lost from the cofactor.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asn255(257)A | metal ligand |
| Asp297(299)A | metal ligand |
| Glu171(173)A | metal ligand |
Chemical Components
electron transfer, native state of cofactor regenerated
Step 7. In an inferred return step, Glu171 is deprotonated by a water molecule.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asn255(257)A | metal ligand |
| Asp297(299)A | metal ligand |
| Glu171(173)A | metal ligand, proton donor |
Chemical Components
inferred reaction step, native state of enzyme regenerated, proton transferIntroduction
In this original addition/elimination mechanism proposal, Asp297 is the only general acid/base. In this mechanism Asp297 abstracts a proton from the alcohol substrate, which initiates a nucleophilic attack on the PQQ cofcator. The cofactor then undergoes an intramolecular elimination to produce the formic acid product. PQQ is then regenerated through two successive single electron transfers from PQQ to cytochrome cL. However, the exact route that the electrons and protons take out of the active site to the heme of the bound cytochrome cL during the reduction of PQQ is unknown (due to lack of crystal structure with the cytochrome bound). It is assumed that the electrons will take the shortest route and may be the conserved disulfide ring, Asp105 and Asn52 and water molecules. The protons from the quinol are thought to be released to the periplasm through a hydrogen bonded network of proton donor and acceptor groups. This is thought to involve Asp303, Arg331. Glu177 and a channel of water molecules leading out of the active site [PMID:12686102].
Catalytic Residues Roles
| UniProt | PDB* (1g72) | ||
| Asp299 | Asp297(299)A | Acts as a general acid/base. | metal ligand, proton acceptor, proton donor |
| Asn257, Glu173 | Asn255(257)A, Glu171(173)A | Forms part of the calcium binding site. | metal ligand |
Chemical Components
cofactor used, intermediate formation, overall reactant used, bimolecular nucleophilic addition, proton transfer, intramolecular elimination, electron transferReferences
- Xia ZX et al. (1999), Biochemistry, 38, 1214-1220. Detailed Active Site Configuration of a New Crystal Form of Methanol Dehydrogenase fromMethylophilusW3A1 at 1.9 Å Resolution†,‡. DOI:10.1021/bi9822574. PMID:9930981.
- Zhang X et al. (2007), Proc Natl Acad Sci U S A, 104, 745-749. Mechanism of methanol oxidation by quinoprotein methanol dehydrogenase. DOI:10.1073/pnas.0610126104. PMID:17215371.
- Anthony C et al. (1994), Biochem J, 304, 665-674. The structure and function of methanol dehydrogenase and related quinoproteins containing pyrrolo-quinoline quinone. DOI:10.1042/bj3040665. PMID:7818466.
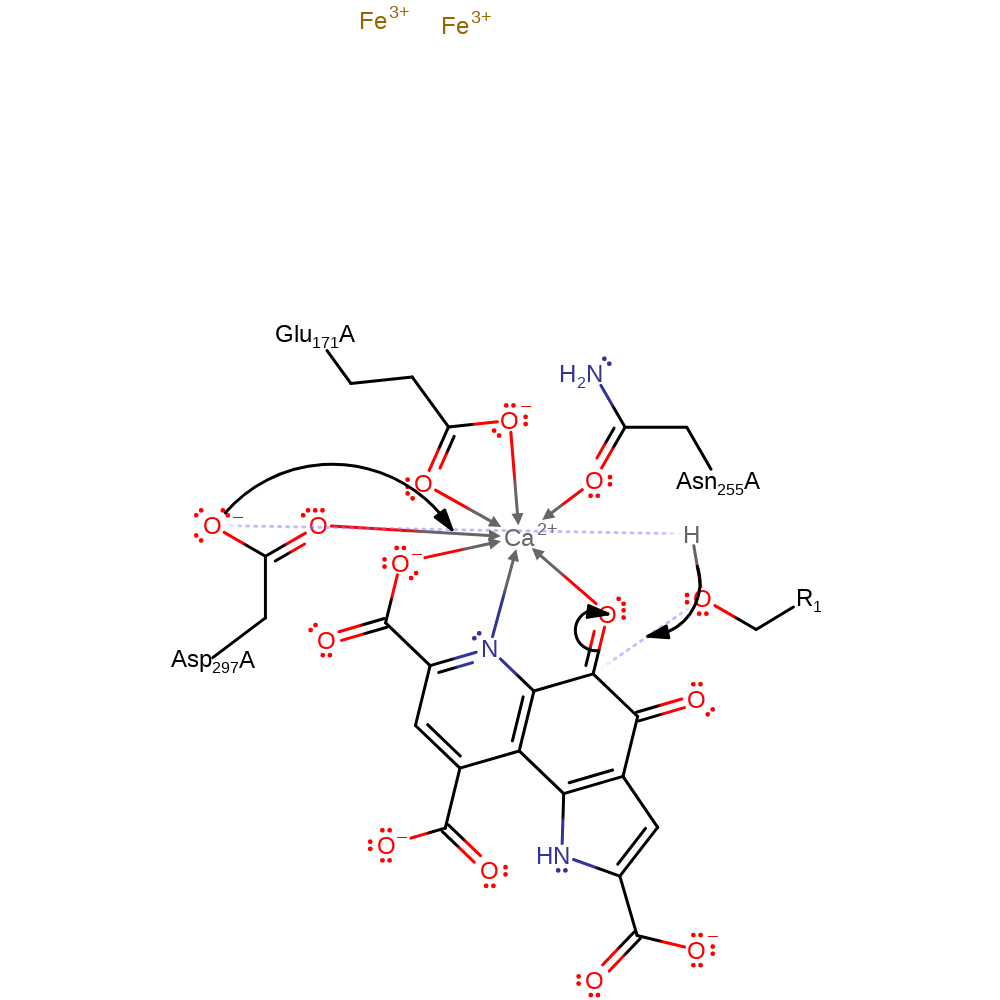
Step 1. Asp297 deprotonates the alcohol group of the primary alcohol, which initiates a nucleophilic attack on the C5 of PQQ.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asn255(257)A | metal ligand |
| Asp297(299)A | metal ligand |
| Glu171(173)A | metal ligand |
| Asp297(299)A | proton acceptor |
Chemical Components
cofactor used, intermediate formation, overall reactant used, ingold: bimolecular nucleophilic addition, proton transferCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asn255(257)A | metal ligand |
| Asp297(299)A | metal ligand |
| Glu171(173)A | metal ligand |
| Asp297(299)A | proton donor |
Chemical Components
proton transfer
Step 3. The oxygen of the PQQ C6 carbonyl group abstracts a proton from the bound primary alcohol, eliminating the reduced PQQ and forming the aldehyde product.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asn255(257)A | metal ligand |
| Asp297(299)A | metal ligand |
| Glu171(173)A | metal ligand |
Chemical Components
ingold: intramolecular elimination, proton transfer
Step 4. In the first step of the reduction of PQQ, a single electron and single proton are lost from the cofactor.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asn255(257)A | metal ligand |
| Asp297(299)A | metal ligand |
| Glu171(173)A | metal ligand |
Chemical Components
electron transfer
Step 5. In the final step of the reduction of PQQ, a single electron and single proton are lost from the cofactor.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asn255(257)A | metal ligand |
| Asp297(299)A | metal ligand |
| Glu171(173)A | metal ligand |





 Download:
Download: 
 Download:
Download: