Oxalate oxidase
Oxalate oxidase (OXO) catalyses the conversion of oxalate and dioxygen to hydrogen peroxide and carbon dioxide. The enzyme is widespread in nature but is most abundant in higher plant tissues, particularly germinating seeds. It is a member of a functionally diverse superfamily known at the cupins or double stranded β-helix proteins containing a mononuclear manganese centre in each subunit.
The mechanism for OXO remains unclear. It is closely related to the bicupin oxalate decarboxylase which also contains manganese but produces carbon dioxide and formate.
Reference Protein and Structure
- Sequence
-
P45850
 (1.2.3.4)
(1.2.3.4)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Hordeum vulgare (Barley)

- PDB
-
2et1
- Oxalate oxidase in complex with substrate analogue glycolate
(1.6 Å)



- Catalytic CATH Domains
-
2.60.120.10
 (see all for 2et1)
(see all for 2et1)
- Cofactors
- Manganese(2+) (1)
Enzyme Reaction (EC:1.2.3.4)
Enzyme Mechanism
Introduction
This mechanism begins with octahedral coordination of the manganese ion at the catalytic centre of the enzyme by three histidine residues (His88, His90 and His137), one glutamate (Glu95) and two solvent water molecules. Monodentate coordination of the oxalate substrate displaces a water molecule from the Mn(II) centre. Then dioxygen binding to Mn(II) displaces another solvent molecule, and forms an Mn(III) superoxide metalloradical complex. The superoxide radical performs a nucleophilic attack on the proximal carboxyl group, activating the substrate. Hydrogen atom transfer forms a distal carboxyl radical species, and the substrate rearranges by homolytic C-C bond cleavage and reduction of Mn(III). Lastly, it is possible that release of products includes hydrolysis of percarbonate (not shown).
Catalytic Residues Roles
| UniProt | PDB* (2et1) | ||
| Asn75 | Asn75A | Asn75 shows conformational flexibility, suggesting it has a dynamic role in catalysis e.g. assisting substrates and products through the access channel. Mutation to alanine results in complete loss of activity. | hydrogen bond acceptor |
| Asn85 | Asn85A | Along with Gln139, Asn85 facilitates formation of the percarbonate product by ensuring the planarity of the manganese ion and atoms of the dioxygen superoxide and the carbon dioxide fragment from oxalate. Mutation to alanine results in complete loss of activity. | hydrogen bond donor |
| Gln139 | Gln139A | A manganese ion, together with Asn75, Asn85 and Gln139 comprise a redox active metal cofactor and hydrogen bonding framework important in the orientation and activation of substrates. | hydrogen bond acceptor, hydrogen bond donor |
| His90, His88, His137, Glu95 | His90A, His88A, His137A, Glu95A | There is octahedral coordination around the manganese ion by His88, His90, Glu95, His137 and two water molecules. | metal ligand |
| Met149, Val77 | Met149A, Val77A | Leu77 and Met149 provide steric constraints on bidentate coordination in OXO. | steric role |
Chemical Components
cofactor used, coordination to a metal ion, decoordination from a metal ion, substitution (not covered by the Ingold mechanisms), radical propagation, colligation, intermediate formation, bimolecular nucleophilic addition, overall reactant used, hydrogen transfer, homolysis, radical termination, intermediate collapse, decarboxylation, overall product formed, native state of cofactor regenerated, inferred reaction step, intramolecular eliminationReferences
- Opaleye O et al. (2006), J Biol Chem, 281, 6428-6433. Structural and spectroscopic studies shed light on the mechanism of oxalate oxidase. DOI:10.1074/jbc.M510256200. PMID:16291738.
- Goodwin JM et al. (2017), PLoS One, 12, e0177164-. Hydrogen peroxide inhibition of bicupin oxalate oxidase. DOI:10.1371/journal.pone.0177164. PMID:28486485.
- Woo EJ et al. (2000), Nat Struct Biol, 7, 1036-1040. Germin is a manganese containing homohexamer with oxalate oxidase and superoxide dismutase activities. DOI:10.1038/80954. PMID:11062559.

Step 1. Monodentate coordination of oxalate monoanion, displacing one solvent molecule from the Mn(II) centre. This forms the dioxygen binding site and lowers the redox potential of the Mn(II)-Mn(III) couple, activating the metal ion to bind dioxygen as a superoxide anion. Leu77 and Met149 provide steric constraints on bidentate coordination.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asn85A | hydrogen bond donor |
| Gln139A | hydrogen bond donor |
| Asn75A | hydrogen bond acceptor |
| Gln139A | hydrogen bond acceptor |
| His90A | metal ligand |
| His88A | metal ligand |
| His137A | metal ligand |
| Glu95A | metal ligand |
| Met149A | steric role |
| Val77A | steric role |
Chemical Components
cofactor used, coordination to a metal ion, decoordination from a metal ion
Step 2. Dioxygen binding to Mn(II) displaces another solvent molecule, and forms an Mn(III) superoxide metalloradical complex.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His90A | metal ligand |
| His88A | metal ligand |
| His137A | metal ligand |
| Glu95A | metal ligand |
| Asn85A | hydrogen bond donor |
| Gln139A | hydrogen bond donor |
| Asn75A | hydrogen bond acceptor |
| Gln139A | hydrogen bond acceptor |
Chemical Components
substitution (not covered by the Ingold mechanisms), coordination to a metal ion, decoordination from a metal ion, cofactor used, radical propagation, colligation, intermediate formation
Step 3. The superoxide radical performs a nucleophilic attack on the proximal carboxyl group, activating the substrate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His90A | metal ligand |
| His88A | metal ligand |
| His137A | metal ligand |
| Glu95A | metal ligand |
| Asn75A | hydrogen bond acceptor |
| Gln139A | hydrogen bond acceptor |
| Asn85A | hydrogen bond donor |
| Gln139A | hydrogen bond donor |
Chemical Components
ingold: bimolecular nucleophilic addition, cofactor used, intermediate formation, radical propagation, overall reactant used
Step 4. Hydrogen atom transfer forms a distal carboxyl radical species.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asn75A | hydrogen bond acceptor |
| Gln139A | hydrogen bond acceptor |
| Asn85A | hydrogen bond donor |
| Gln139A | hydrogen bond donor |
| His90A | metal ligand |
| His88A | metal ligand |
| His137A | metal ligand |
| Glu95A | metal ligand |
Chemical Components
hydrogen transfer, radical propagation
Step 5. Substrate radical rearranges with homolytic C-C bond cleavage and reduction of Mn(III) to Mn(II).
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asn85A | hydrogen bond donor |
| Gln139A | hydrogen bond donor |
| Asn75A | hydrogen bond acceptor |
| Gln139A | hydrogen bond acceptor |
| His90A | metal ligand |
| His88A | metal ligand |
| His137A | metal ligand |
| Glu95A | metal ligand |
Chemical Components
homolysis, radical termination, intermediate collapse, decarboxylation, decoordination from a metal ion
Step 6. The remaining products are formed by an intramolecular elimination. Two water molecules bind the metal to regenerate the initial step. This step is inferred by the curator.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asn75A | hydrogen bond acceptor |
| Gln139A | hydrogen bond acceptor |
| Asn85A | hydrogen bond donor |
| Gln139A | hydrogen bond donor |
| His90A | metal ligand |
| His88A | metal ligand |
| His137A | metal ligand |
| Glu95A | metal ligand |
Chemical Components
overall product formed, native state of cofactor regenerated, inferred reaction step, ingold: intramolecular eliminationIntroduction
This mechanism identifies the fraction of the enzyme containing Mn(III) as the active form of the enzyme. Monodentate coordination of the oxalate monoanion to an Mn(III) centre. Under anaerobic conditions, oxalate reduces Mn(III) to Mn(II) and forms an oxalyl free radical. Decarboxylation leaves a carbon dioxide radical anion bound to Mn(II). Dioxygen intercepts the carbon dioxide radical anion, generating a second molecule of carbon dioxide and superoxide. Subsequent electron transfer from Mn(II) to a hydroperoxyl radical generates hydrogen peroxide. This mechanism may be specific to barley oxalate oxidase.
Catalytic Residues Roles
| UniProt | PDB* (2et1) | ||
| Asn85 | Asn85A | Along with Gln139, Asn85 facilitates formation of the percarbonate product by ensuring the planarity of the manganese ion and atoms of the dioxygen superoxide and the carbon dioxide fragment from oxalate. Mutation to alanine results in complete loss of activity. | hydrogen bond donor |
| Asn75, Asn85, Gln139 | Asn75A, Asn85A, Gln139A | A manganese ion, together with Asn75, Asn85 and Gln139 comprise a redox active metal cofactor and hydrogen bonding framework important in the orientation and activation of substrates. | hydrogen bond acceptor |
| His90, His88, His137, Glu95 | His90A, His88A, His137A, Glu95A | There is octahedral coordination around the manganese ion by His88, His90, Glu95, His137 and two water molecules. | metal ligand |
| Met149, Val77 | Met149A, Val77A | Leu77 and Met149 provide steric constraints on bidentate coordination in OXO. | steric role |
Chemical Components
substitution (not covered by the Ingold mechanisms), decoordination from a metal ion, coordination to a metal ion, cofactor used, intermediate formation, proton transfer, decarboxylation, intermediate collapse, electron transfer, overall product formed, radical termination, native state of cofactor regeneratedReferences
- Whittaker MM et al. (2007), J Biol Chem, 282, 7011-7023. Burst kinetics and redox transformations of the active site manganese ion in oxalate oxidase: implications for the catalytic mechanism. DOI:10.1074/jbc.M609374200. PMID:17210574.

Step 1. Monodentate coordination of oxalate monoanion, displacing one solvent molecule from an Mn(III) centre.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Val77A | steric role |
| Met149A | steric role |
| Glu95A | metal ligand |
| His137A | metal ligand |
| His88A | metal ligand |
| His90A | metal ligand |
| Gln139A | hydrogen bond acceptor |
| Asn75A | hydrogen bond acceptor |
| Gln139A | hydrogen bond donor |
| Asn85A | hydrogen bond donor |
Chemical Components
substitution (not covered by the Ingold mechanisms), decoordination from a metal ion, coordination to a metal ion, cofactor used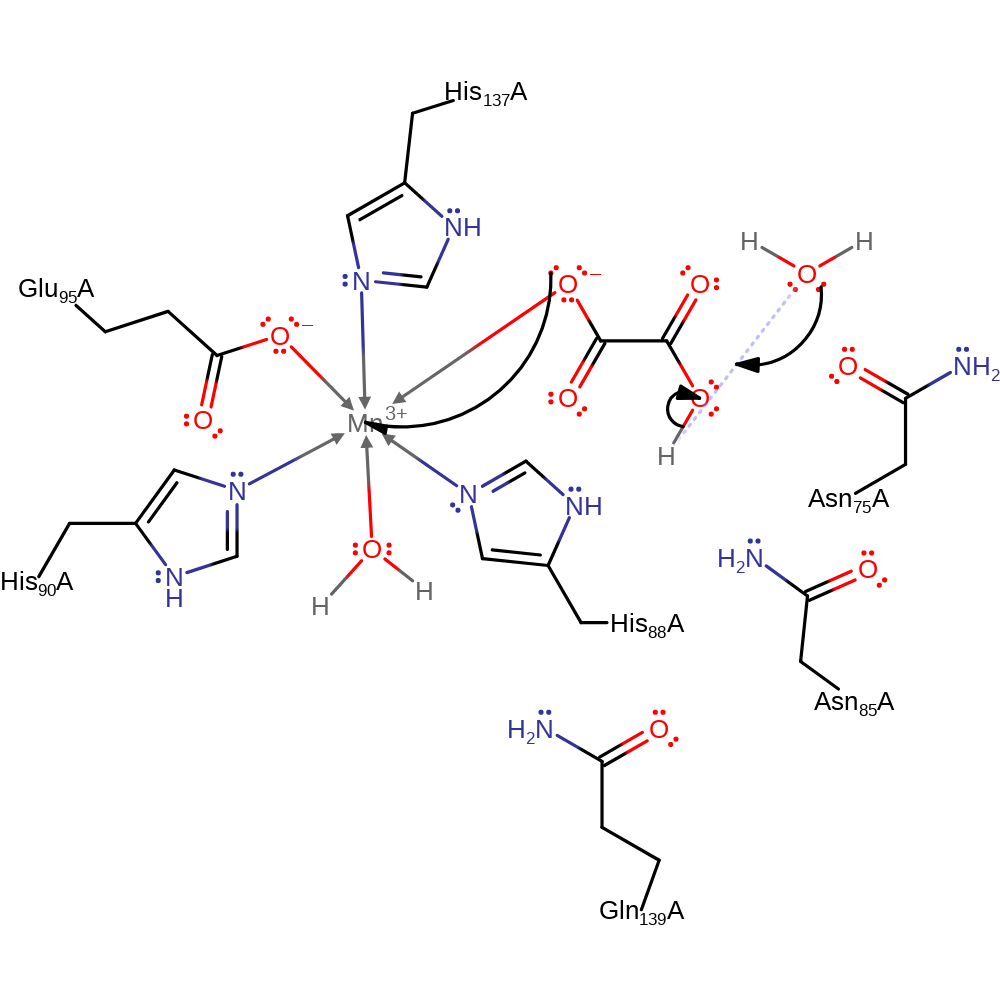
Step 2. Under anaerobic conditions, oxalate reduces Mn(III) to Mn(II) and forms an oxalyl free radical. Proton transfer inferred by curator.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Gln139A | hydrogen bond acceptor |
| Asn75A | hydrogen bond acceptor |
| Gln139A | hydrogen bond donor |
| Asn85A | hydrogen bond donor |
| Glu95A | metal ligand |
| His137A | metal ligand |
| His88A | metal ligand |
| His90A | metal ligand |
Chemical Components
intermediate formation, cofactor used, decoordination from a metal ion, coordination to a metal ion, substitution (not covered by the Ingold mechanisms), proton transfer
Step 3. Decarboxylation in aqueous conditions releases carbon dioxide and leaves a carbon dioxide radical anion bound to Mn(II).
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
decarboxylation, intermediate collapse, decoordination from a metal ion
Step 4. Dioxygen intercepts the carbon dioxide radical anion, generating a second molecule of carbon dioxide and superoxide. Protonation inferred by curator.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
decarboxylation, decoordination from a metal ion, electron transfer
Step 5. Electron transfer from Mn(II) to a hydroperoxyl radical, generating hydrogen peroxide. A other molecule binds the metal ion to regenerate the active site.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
electron transfer, overall product formed, proton transfer, radical termination, native state of cofactor regeneratedIntroduction
This mechanism is consistent with the results from a study demonstrating that oxalate oxidase and oxalate decarboxylase activities can be interchanged by mutating an active site lid. Oxalate binds to Mn(II), which then undergoes oxidation to Mn(III) upon binding of dioxygen. Reduction of Mn(III) to Mn(II) and decarboxylation occur, which is followed by insertion of the superoxide radical into oxalate. If the insertion occurred before decarboxylation, the mechanism would be committed to oxidation.
Catalytic Residues Roles
| UniProt | PDB* (2et1) | ||
| Asn85 | Asn85A | Along with Gln139, Asn85 facilitates formation of the percarbonate product by ensuring the planarity of the manganese ion and atoms of the dioxygen superoxide and the carbon dioxide fragment from oxalate. Mutation to alanine results in complete loss of activity. | hydrogen bond donor |
| Asn75, Asn85, Gln139 | Asn75A, Asn85A, Gln139A | A manganese ion, together with Asn75, Asn85 and Gln139 comprise a redox active metal cofactor and hydrogen bonding framework important in the orientation and activation of substrates. | hydrogen bond acceptor |
| His90, His88, His137, Glu95 | His90A, His88A, His137A, Glu95A | There is octahedral coordination around the manganese ion by His88, His90, Glu95, His137 and two water molecules. | metal ligand |
| Met149, Val77 | Met149A, Val77A | Leu77 and Met149 provide steric constraints on bidentate coordination in OXO. | steric role |
Chemical Components
substitution (not covered by the Ingold mechanisms), decoordination from a metal ion, coordination to a metal ion, cofactor used, proton transfer, electron transfer, decarboxylation, colligation, intermediate formation, overall product formedReferences
- Burrell MR et al. (2007), Biochemistry, 46, 12327-12336. Oxalate decarboxylase and oxalate oxidase activities can be interchanged with a specificity switch of up to 282,000 by mutating an active site lid. DOI:10.1021/bi700947s. PMID:17924657.
- Zhu W et al. (2016), Biochemistry, 55, 2163-2173. Substrate Binding Mode and Molecular Basis of a Specificity Switch in Oxalate Decarboxylase. DOI:10.1021/acs.biochem.6b00043. PMID:27014926.

Step 1. Monodentate coordination of oxalate monoanion, displacing one solvent molecule from the Mn(II) centre.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Val77A | steric role |
| Met149A | steric role |
| Glu95A | metal ligand |
| His137A | metal ligand |
| His88A | metal ligand |
| His90A | metal ligand |
| Gln139A | hydrogen bond acceptor |
| Asn75A | hydrogen bond acceptor |
| Gln139A | hydrogen bond donor |
| Asn85A | hydrogen bond donor |
Chemical Components
substitution (not covered by the Ingold mechanisms), decoordination from a metal ion, coordination to a metal ion, cofactor used
Step 2. Dioxygen binding to Mn(II) displaces another solvent molecule, and forms an Mn(III) superoxide metalloradical complex.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Gln139A | hydrogen bond acceptor |
| Asn75A | hydrogen bond acceptor |
| Gln139A | hydrogen bond donor |
| Asn85A | hydrogen bond donor |
| Glu95A | metal ligand |
| His137A | metal ligand |
| His88A | metal ligand |
| His90A | metal ligand |
Chemical Components
cofactor used, decoordination from a metal ion, coordination to a metal ion, substitution (not covered by the Ingold mechanisms)
Step 3. Activation of the Mn(III)-bound oxalate through deprotonation (inferred by curator). This causes the oxalate bridging oxygen to transfer an electron to the Mn(III) centre.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Gln139A | hydrogen bond donor |
| Asn85A | hydrogen bond donor |
| Gln139A | hydrogen bond acceptor |
| Asn75A | hydrogen bond acceptor |
| Glu95A | metal ligand |
| His137A | metal ligand |
| His88A | metal ligand |
| His90A | metal ligand |
Chemical Components
cofactor used, proton transfer, electron transferCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
decarboxylation
Step 5. The superoxide radical binds to the the radical carbonyl group.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
colligation, intermediate formation
Step 6. A second decarboxylation occurs and along with two proton transfers, to form hydrogen peroxide.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|





 Download:
Download:  Download:
Download: 
 Download:
Download: