- Course overview
- Search within this course
- Introduction
- Real-time PCR
- Microarrays
- What is Next Generation DNA Sequencing?
- RNA sequencing
- Biological interpretation of gene expression data
- Genotyping, epigenetic and DNA/RNA-protein interaction methods
- DNA/RNA-protein interactions
- Summary
- Quiz: Check your learning
- Your feedback
- Learn more
- References
Real-time PCR
Real-time polymerase chain reaction (real-time PCR) is commonly used to measure gene expression. It is more sensitive than microarrays in detecting small changes in expression but requires more input RNA and is less adaptable to high-throughput studies (1). It is best suited for studies of small subsets of genes. Its one major shortcoming is that the sequence of the specific target gene of interest must be known (so you can design the PCR primers), hence real-time PCR can only be used for studying known genes.
Real-time PCR steps
The first step in a real-time PCR reaction is the conversion of RNA to complementary DNA (cDNA) – this process is known as reverse transcription (Figure 1). The next step uses fluorescent reporters and a PCR reaction to amplify and detect specific genes (Figure 1). Two types of fluorescent reporters are commonly used; these are SYBR green and Taqman probes.
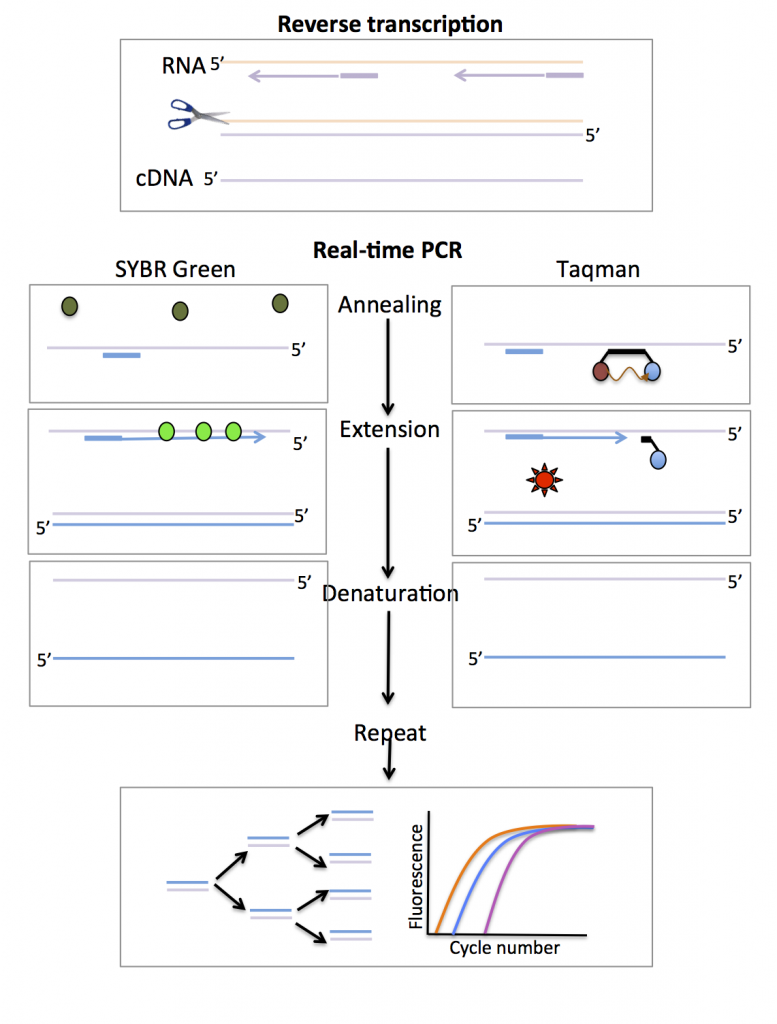
SYBR green and Taqman probes
SYBR green is a dye that fluoresces only when bound to double stranded DNA (i.e the PCR product) (Figure 1).
Taqman probes are made of a gene-specific nucleic acid probe, joined to reporter and quencher molecules. The probe binds to the DNA between the forward and reverse primer. While the reporter and quencher are bound to the probe, the quencher absorbs the fluorescence emitted by the reporter. During the extension phase of the PCR reaction the probe is degraded, releasing the reporter and allowing its fluorescence to be detected. The advantage of the Taqman method is that probes with different coloured reporters can be combined in multiplex assays.
For both SYBR green and Taqman methods, the amount of fluorescence in a sample is detected in ‘real-time’ and plotted against the cycle number (Figure 1). The amount of fluorescence is proportional to the amount of PCR product. The time point at which the fluorescence reaches a defined threshold is relative to the level of gene expression. The design of real-time PCR experiments requires prior knowledge of the gene sequence and careful consideration of the types of controls to include.
|
|