- Course overview
- Search within this course
- Introduction
- Real-time PCR
- Microarrays
- What is Next Generation DNA Sequencing?
- Biological interpretation of gene expression data
- Genotyping, epigenetic and DNA/RNA-protein interaction methods
- DNA/RNA-protein interactions
- Summary
- Quiz: Check your learning
- Your feedback
- Learn more
- References
Differential gene expression analysis
Differential expression analysis means taking the normalised read count data and performing statistical analysis to discover quantitative changes in expression levels between experimental groups. For example, we use statistical testing to decide whether, for a given gene, an observed difference in read counts is significant, that is, whether it is greater than what would be expected just due to natural random variation.
Methods for differential expression analysis
There are different methods for differential expression analysis such as edgeR or DESeq2 are based on negative binomial (NB) distributions or baySeq and EBSeq which are Bayesian approaches based on a negative binomial model. It is important to consider the experimental design when choosing an analysis method. While some of the differential expression tools can only perform pair-wise comparison, others such as edgeR, limma-voom, DESeq2 and maSigPro can perform multiple comparisons.
In Figure 16, below, we outline the RNA-seq processing pipeline used to generate data for Expression Atlas.
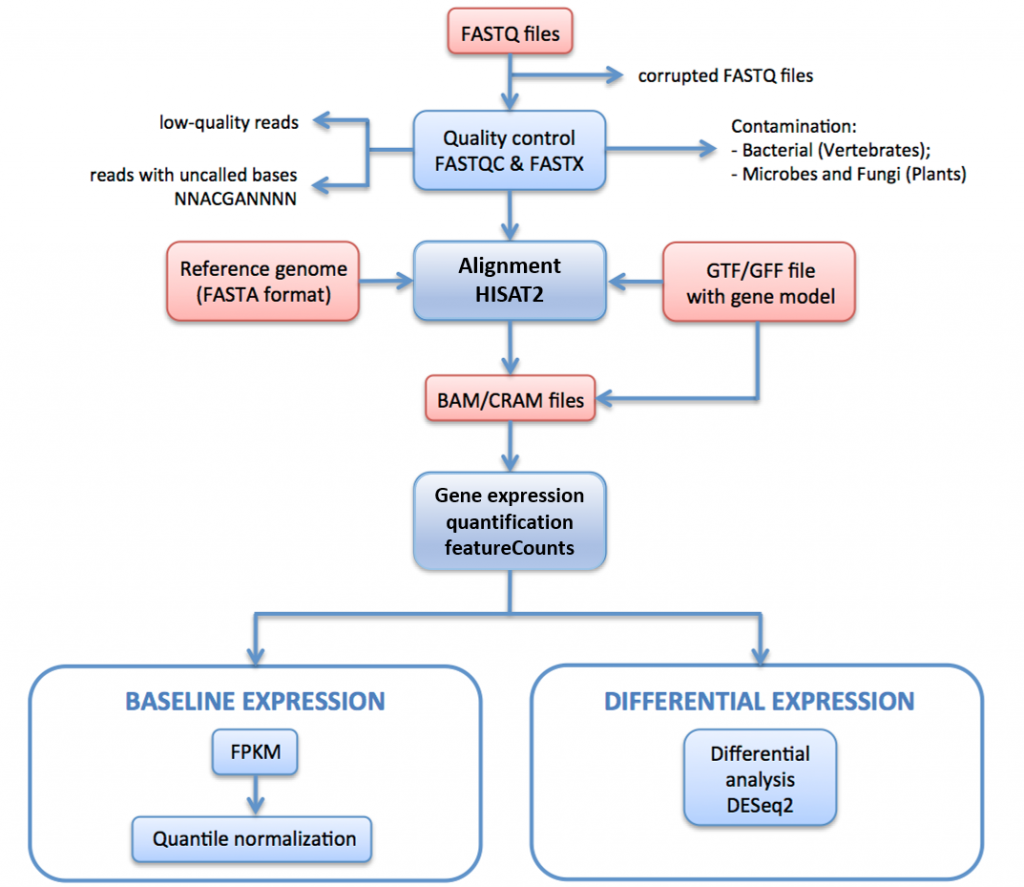
In this pipeline raw reads (FASTQ files) undergo quality assessment and filtering. The quality-filtered reads are aligned to the reference genome via HISAT2. The mapped reads are summarised and aggregated over genes via HTSeq. For baseline expression, the FPKMs are calculated from the raw counts by iRAP. These are averaged for each set of technical replicates, and then quantile normalised within each set of biological replicates using limma.
Finally, they are averaged for all biological replicates (if any). For differential expression, genes expressed differentially between the test and the reference groups of each pairwise contrast are identified using DESeq2.