- Course overview
- Search within this course
- Network analysis in biology
- Introduction to graph theory
- Types of biological networks
- The sources of data underlying biological networks
- Building and analysing PPINs
- Summary
- Quiz: Check your learning
- Your feedback
- Learn more
- References
Protein-protein interaction networks
Protein-protein interactions (PPIs) are essential to almost every process in a cell, so understanding PPIs is crucial for understanding cell physiology in normal and disease states. It is also essential in drug development, since drugs can affect PPIs. Protein-protein interaction networks (PPIN) are mathematical representations of the physical contacts between proteins in the cell. These contacts:
- are specific
- occur between defined binding regions in the proteins
- have a particular biological meaning (i.e., they serve a specific function)
PPI information can represent both transient and stable interactions:
- Stable interactions are formed in protein complexes (e.g. ribosome, haemoglobin)
- Transient interactions are brief interactions that modify or carry a protein, leading to further change (e.g. protein kinases, nuclear pore importins). They constitute the most dynamic part of the interactome
Knowledge of PPIs can be used to:
- assign putative roles to uncharacterised proteins
- add fine-grained detail about the steps within a signalling pathway
- characterise the relationships between proteins that form multi-molecular complexes such as the proteasome
The interactome
The interactome is the totality of PPIs that happen in a cell, an organism or a specific biological context. The development of large-scale PPI screening techniques, especially high-throughput affinity purification combined with mass-spectrometry and the yeast two-hybrid assay, has caused an explosion in the amount of PPI data and the construction of ever more complex and complete interactomes (Figure 16). This experimental evidence is complemented by the availability of PPI prediction algorithms. A lot of this information is available through molecular interaction databases such as IntAct.
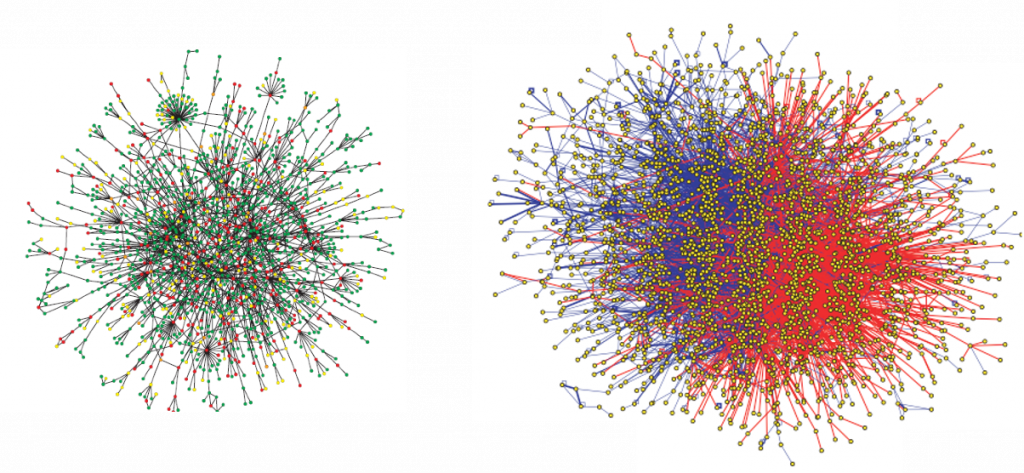
It is important to emphasise once more the limitations of available PPI data. Our current knowledge of the interactome is both incomplete and noisy. PPI detection methods have limitations as to how many truly physiological interactions they can detect and they all find false positives and negatives.
On the next few pages we will take a look at some of the properties of protein-protein interaction networks and the implications of these properties for biology.