- Course overview
- Search within this course
- What is UniProt?
- Where does the data come from?
- Why do we need UniProt?
- When to use UniProt
- Quiz: Check your learning I
- How to access and navigate UniProt
- How to search UniProt
- Annotation score
- Quiz: Check your learning II
- How to use UniProt tools
- How to get data from UniProt
- How to submit data to UniProt
- When to use UniProt: guided example
- Exercise: finding entries with 3D structures
- Exercise: mapping other database identifiers to UniProt
- Summary
- Your feedback
- Get help and support on UniProt
- References
Exploring the Protein Feature Viewer
Data visualisation and integration have become increasingly important as biological data continues to grow at exponential rates. The protein feature viewer uses a highly interactive BioJS component which allows you to see different types of protein sequence features such as domains, sites, post-translational modifications, topology, mutagenesis, proteomics-derived peptides, antigenic sequences and natural variants from multiple sources in a single view. You have been looking at aspects of this information in the relevant sections of the protein entry, the Feature Viewer tab in each protein entry allows you to bring all this graphical information together in a single view and use it to understand different aspects of protein function.
Similar to genome viewers, the UniProt feature viewer displays information on tracks. Each track presents a biological category of features, and can be expanded into sub-tracks for a more detailed overview. When an experimentally derived 3D structure is available, this is shown below and the feature selected is also highlighted on this.

Keep it in sight
Showing all co-localised sequence features in one view makes it easier to spot patterns and make inferences. Figure 32 shows an example of how you might use the feature viewer. To investigate the involvement of human lipoprotein lipase (UniProtKB entry P06858) in lipoprotein lipase deficiency, a rare autosomal recessive lipid disorder, you can click on the “Domain & sites” label and see that there are three active sites (Figure 33).
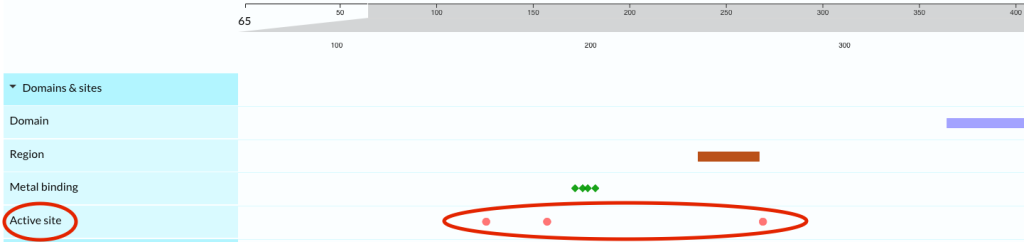
If you want to investigate the potential effect of variants on these sites, click on the “Variants” label to open up the track. Clicking on the second active site at position 183 highlights three disease variants in the same position. You can then zoom into the active site and click on this to see the corresponding positions highlighted across all protein features (Figure 34). In this case, point mutations created at the same residue confirm the variant effect is most probably due to a loss of enzyme function. Clicking on the variant dots opens up an information box with links to relevant publications. This unique functionality lets you view possible variant implications at a glance. Click on ![]() to view the information, about Active site, mutagenesis and variants, that you would see for the position 183.
to view the information, about Active site, mutagenesis and variants, that you would see for the position 183.
Figure 34 Feature Viewer example highlighting natural variants colocated with an active site.
Want to integrate the feature viewer into your website?
The UniProt feature viewer can be integrated into any website. You can choose to keep all available tracks or only those most relevant to you.
If you would like to include the feature viewer in your own website or resource, you can find instructions in our technical documentation here.