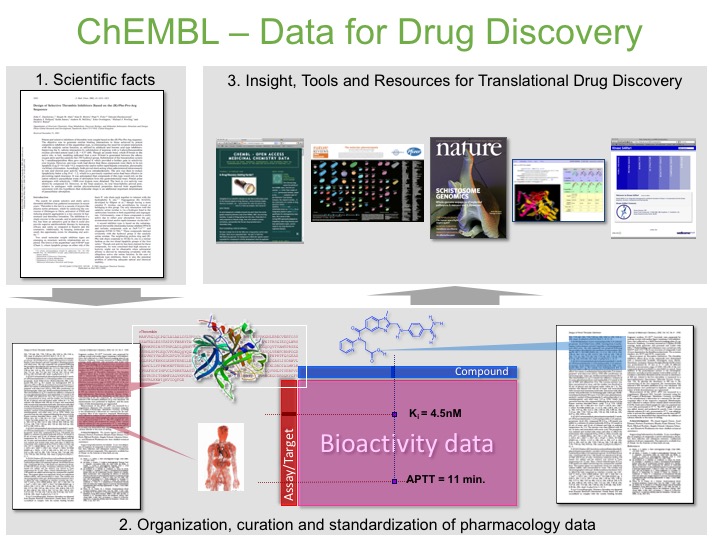
How is ChEMBL data curated?
We identify scientific facts in a journal article and then extract the information and add it to the ChEMBL database in a structured format (Figure 3).
For example an article might have structure activity relationship (SAR) data for a series of related compounds binding to a specific protein in the form of a number of IC50 values. In this case we will draw the chemical structures as molfiles or smiles. We will identify the Uniprot accession number for the protein and the organism it is from and extract a short description of the experiment (or assay as we call it) that the IC50 values were measured in. This information is then added to the ChEMBL database. You can then search it for specific target by name or sequence or do substructure or similarity searches to find data on specific chemotypes.
If an article has data on measurements made in cell-based assays, in vivo pharmacology assays or pharmacokinetic studies for compounds we will also extract that and add it to the ChEMBL database.
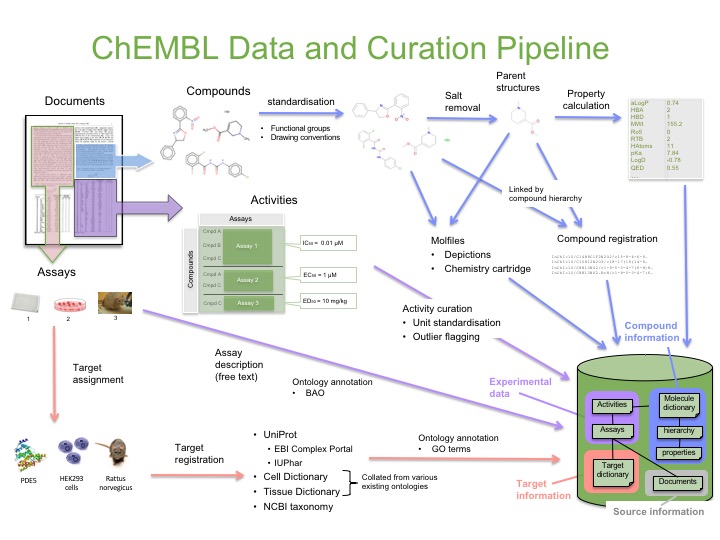
We then perform additional curation and annotation of the data so that we can provide a high quality database for you to use in your research. This curation process is shown in the figure below.