What is functional genomics?
Functional genomics is the study of how genes and intergenic regions of the genome contribute to different biological processes. A researcher in this field typically studies genes or regions on a “genome-wide” scale (i.e. all or multiple genes/regions at the same time), with the hope of narrowing them down to a list of candidate genes or regions to analyse in more detail.
The goal of functional genomics is to determine how the individual components of a biological system work together to produce a particular phenotype. Functional genomics focuses on the dynamic expression of gene products in a specific context, for example, at a specific developmental stage or during a disease. In functional genomics, we try to use our current knowledge of gene function to develop a model linking genotype to phenotype.
There are several specific functional genomics approaches depending on what we are focused on (Figure 2):
- DNA level (genomics and epigenomics)
- RNA level (transcriptomics)
- Protein level (proteomics)
- Metabolite level (metabolomics)
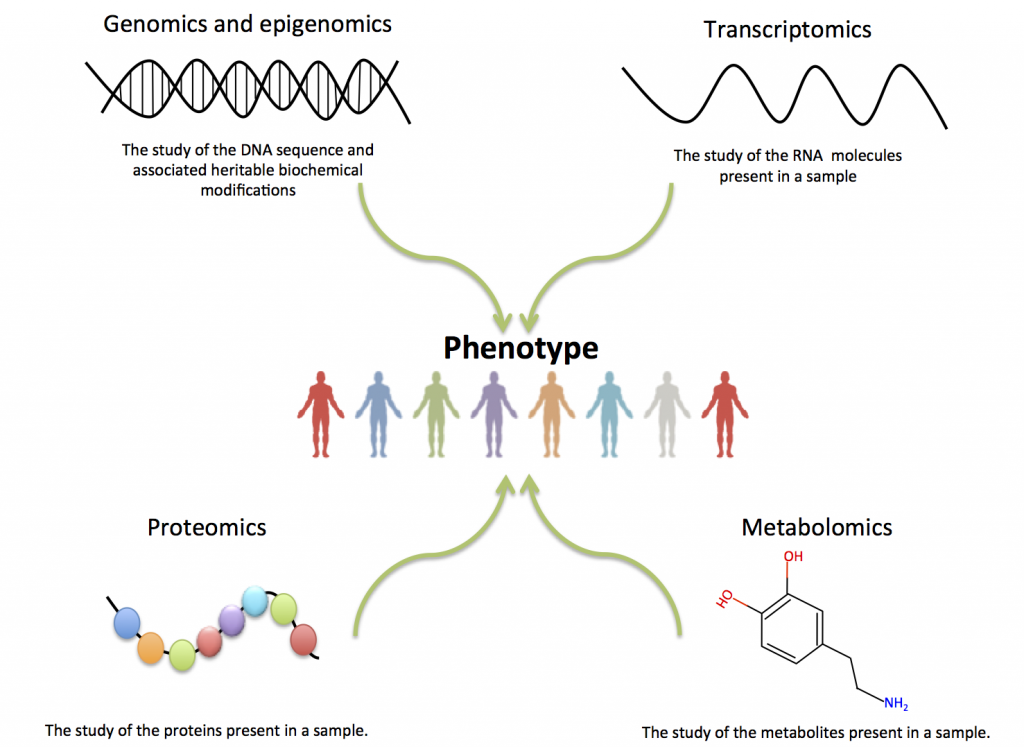
Together, transcriptomics, proteomics and metabolomics describe the transcripts, proteins and metabolites of a biological system, and the integration of these data is expected to provide a complete model of the biological system under study. In you are interested in knowing more about those different approaches and their integration, you can try our curated collection ‘Methods and resources for omics studies’.
In this course, we focus on DNA- and RNA-level approaches.