- Course overview
- Search within this course
- Why do we need a pathway database?
- What is Reactome?
- When to use Reactome
- Who is Reactome for?
- Understanding the Pathway Browser
- Analysing data
- Programmatic access
- Contributing to Reactome
- Summary
- Try it yourself!
- Your feedback
- Get help and support on Reactome
Searching Reactome
Simple text or identifier searches
Global search
You can search the whole of Reactome content by entering your query into the Search box, which is located at the top of the homepage just below the logo and menu bar (boxed with a red dotted line in Figure 12). You can use descriptive text names, many database accessions or identifiers for proteins, compounds and genes, or genes symbols. Ideally, to find a particular protein use its UniProt ID, for compounds use the ChEBI ID. The search is indexed so as you type you may see suggestions. If the item you intend to search for is suggested, select it and the results page will appear. Otherwise, after entering your query, click on the Go! Button.
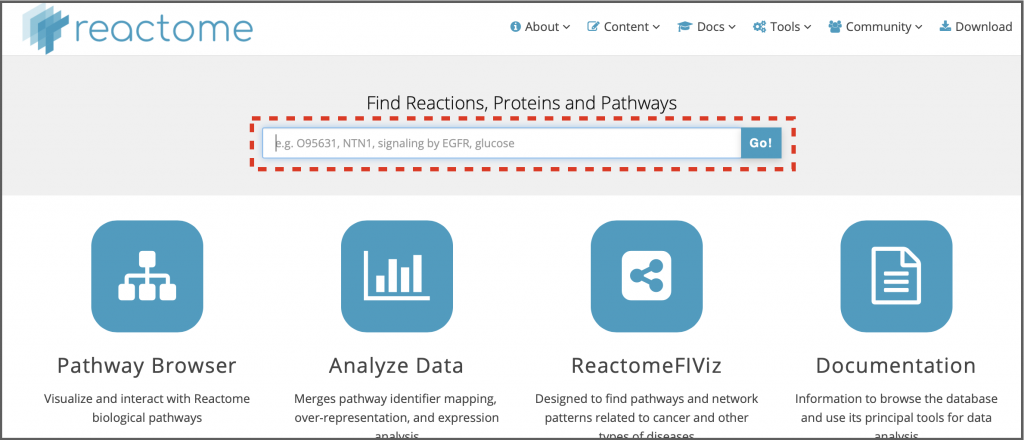
Let’s try an example to search for human EGFR signaling in Reactome.
1. Type ‘EGFR signaling’ into the search box and click Go!
2. The results will appear as shown in Figure 13, with the total number of hits indicated in the top left. Results are sorted into categories, e.g. Reaction, Pathway, Protein, each displayed in its own section, with an indication of the number of hits in that section
3. You can filter the results by selecting the appropriate check boxes from the panel on the left side (red box on Figure 13). Select check boxes for the categories you want to see. Note that Homo sapiens is the default species
4. Click on any of the results to go to the corresponding details page (Figure 15, next page)
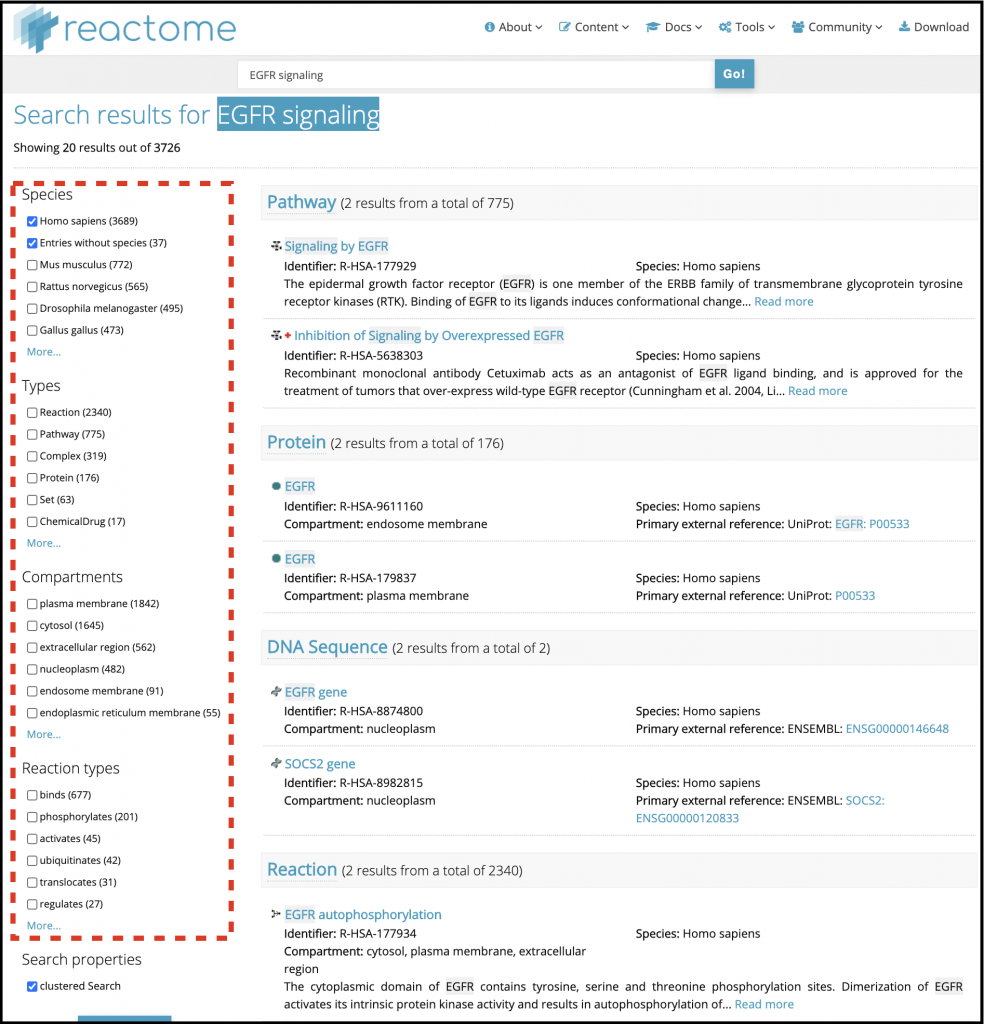
Most results have descriptive text details. Your search terms will be highlighted if they appear within the title or descriptive text.