EMD-12694
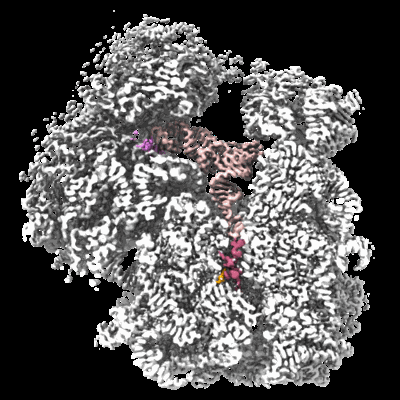
Cryo-EM structure of an Escherichia coli TnaC(R23F)-ribosome complex stalled in response to L-tryptophan
EMD-12694
Single-particle2.4 Å
 Deposition: 29/03/2021
Deposition: 29/03/2021Map released: 01/09/2021
Last modified: 13/11/2024
Sample Organism:
Escherichia coli (strain K12)
Sample: 70S ribosome stalled during translation of TnaC(R23F) with Pro24 in the P-site.
Fitted models: 7o1a (Avg. Q-score: 0.573)
Raw data: EMPIAR-10695
Deposition Authors: van der Stel AX ,
Gordon ER
,
Gordon ER 
Sample: 70S ribosome stalled during translation of TnaC(R23F) with Pro24 in the P-site.
Fitted models: 7o1a (Avg. Q-score: 0.573)
Raw data: EMPIAR-10695
Deposition Authors: van der Stel AX
 ,
Gordon ER
,
Gordon ER 
Structural basis for the tryptophan sensitivity of TnaC-mediated ribosome stalling.
van der Stel AX  ,
Gordon ER
,
Gordon ER  ,
Sengupta A
,
Sengupta A  ,
Martinez AK,
Klepacki D,
Perry TN,
Herrero Del Valle A
,
Martinez AK,
Klepacki D,
Perry TN,
Herrero Del Valle A  ,
Vazquez-Laslop N
,
Vazquez-Laslop N  ,
Sachs MS
,
Sachs MS  ,
Cruz-Vera LR
,
Cruz-Vera LR  ,
Innis CA
,
Innis CA 
(2021) Nat Commun , 12 , 5340 - 5340
 ,
Gordon ER
,
Gordon ER  ,
Sengupta A
,
Sengupta A  ,
Martinez AK,
Klepacki D,
Perry TN,
Herrero Del Valle A
,
Martinez AK,
Klepacki D,
Perry TN,
Herrero Del Valle A  ,
Vazquez-Laslop N
,
Vazquez-Laslop N  ,
Sachs MS
,
Sachs MS  ,
Cruz-Vera LR
,
Cruz-Vera LR  ,
Innis CA
,
Innis CA 
(2021) Nat Commun , 12 , 5340 - 5340
Abstract:
Free L-tryptophan (L-Trp) stalls ribosomes engaged in the synthesis of TnaC, a leader peptide controlling the expression of the Escherichia coli tryptophanase operon. Despite extensive characterization, the molecular mechanism underlying the recognition and response to L-Trp by the TnaC-ribosome complex remains unknown. Here, we use a combined biochemical and structural approach to characterize a TnaC variant (R23F) with greatly enhanced sensitivity for L-Trp. We show that the TnaC-ribosome complex captures a single L-Trp molecule to undergo termination arrest and that nascent TnaC prevents the catalytic GGQ loop of release factor 2 from adopting an active conformation at the peptidyl transferase center. Importantly, the L-Trp binding site is not altered by the R23F mutation, suggesting that the relative rates of L-Trp binding and peptidyl-tRNA cleavage determine the tryptophan sensitivity of each variant. Thus, our study reveals a strategy whereby a nascent peptide assists the ribosome in detecting a small metabolite.
Free L-tryptophan (L-Trp) stalls ribosomes engaged in the synthesis of TnaC, a leader peptide controlling the expression of the Escherichia coli tryptophanase operon. Despite extensive characterization, the molecular mechanism underlying the recognition and response to L-Trp by the TnaC-ribosome complex remains unknown. Here, we use a combined biochemical and structural approach to characterize a TnaC variant (R23F) with greatly enhanced sensitivity for L-Trp. We show that the TnaC-ribosome complex captures a single L-Trp molecule to undergo termination arrest and that nascent TnaC prevents the catalytic GGQ loop of release factor 2 from adopting an active conformation at the peptidyl transferase center. Importantly, the L-Trp binding site is not altered by the R23F mutation, suggesting that the relative rates of L-Trp binding and peptidyl-tRNA cleavage determine the tryptophan sensitivity of each variant. Thus, our study reveals a strategy whereby a nascent peptide assists the ribosome in detecting a small metabolite.
