EMD-23708
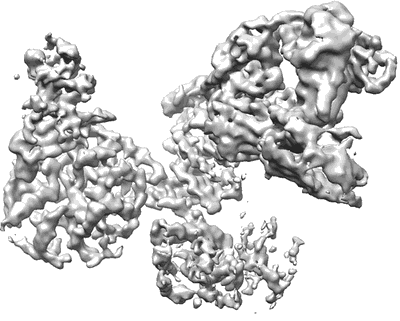
ATP-bound AMP-activated protein kinase
EMD-23708
Single-particle3.93 Å
 Deposition: 26/03/2021
Deposition: 26/03/2021Map released: 15/12/2021
Last modified: 06/11/2024
Sample Organism:
Homo sapiens,
Escherichia coli,
Lama glama
Sample: MBP-fused ATP bound AMPK in complex with C-compound stabilized by Fab and a nanobody
Fitted models: 7m74 (Avg. Q-score: 0.27)
Deposition Authors: Yan Y ,
Mukherjee S
,
Mukherjee S 
Sample: MBP-fused ATP bound AMPK in complex with C-compound stabilized by Fab and a nanobody
Fitted models: 7m74 (Avg. Q-score: 0.27)
Deposition Authors: Yan Y
 ,
Mukherjee S
,
Mukherjee S 
Structure of an AMPK complex in an inactive, ATP-bound state.
Yan Y  ,
Mukherjee S
,
Mukherjee S  ,
Harikumar KG
,
Harikumar KG  ,
Strutzenberg TS
,
Strutzenberg TS  ,
Zhou XE
,
Zhou XE  ,
Suino-Powell K,
Xu TH
,
Suino-Powell K,
Xu TH  ,
Sheldon RD
,
Sheldon RD  ,
Lamp J
,
Lamp J  ,
Brunzelle JS
,
Brunzelle JS  ,
Radziwon K,
Ellis A
,
Radziwon K,
Ellis A  ,
Novick SJ
,
Novick SJ  ,
Vega IE,
Jones RG
,
Vega IE,
Jones RG  ,
Miller LJ
,
Miller LJ  ,
Xu HE
,
Xu HE  ,
Griffin PR
,
Griffin PR  ,
Kossiakoff AA,
Melcher K
,
Kossiakoff AA,
Melcher K 
(2021) Science , 373 , 413 - 419
 ,
Mukherjee S
,
Mukherjee S  ,
Harikumar KG
,
Harikumar KG  ,
Strutzenberg TS
,
Strutzenberg TS  ,
Zhou XE
,
Zhou XE  ,
Suino-Powell K,
Xu TH
,
Suino-Powell K,
Xu TH  ,
Sheldon RD
,
Sheldon RD  ,
Lamp J
,
Lamp J  ,
Brunzelle JS
,
Brunzelle JS  ,
Radziwon K,
Ellis A
,
Radziwon K,
Ellis A  ,
Novick SJ
,
Novick SJ  ,
Vega IE,
Jones RG
,
Vega IE,
Jones RG  ,
Miller LJ
,
Miller LJ  ,
Xu HE
,
Xu HE  ,
Griffin PR
,
Griffin PR  ,
Kossiakoff AA,
Melcher K
,
Kossiakoff AA,
Melcher K 
(2021) Science , 373 , 413 - 419
Abstract:
Adenosine monophosphate (AMP)-activated protein kinase (AMPK) regulates metabolism in response to the cellular energy states. Under energy stress, AMP stabilizes the active AMPK conformation, in which the kinase activation loop (AL) is protected from protein phosphatases, thus keeping the AL in its active, phosphorylated state. At low AMP:ATP (adenosine triphosphate) ratios, ATP inhibits AMPK by increasing AL dynamics and accessibility. We developed conformation-specific antibodies to trap ATP-bound AMPK in a fully inactive, dynamic state and determined its structure at 3.5-angstrom resolution using cryo-electron microscopy. A 180° rotation and 100-angstrom displacement of the kinase domain fully exposes the AL. On the basis of the structure and supporting biophysical data, we propose a multistep mechanism explaining how adenine nucleotides and pharmacological agonists modulate AMPK activity by altering AL phosphorylation and accessibility.
Adenosine monophosphate (AMP)-activated protein kinase (AMPK) regulates metabolism in response to the cellular energy states. Under energy stress, AMP stabilizes the active AMPK conformation, in which the kinase activation loop (AL) is protected from protein phosphatases, thus keeping the AL in its active, phosphorylated state. At low AMP:ATP (adenosine triphosphate) ratios, ATP inhibits AMPK by increasing AL dynamics and accessibility. We developed conformation-specific antibodies to trap ATP-bound AMPK in a fully inactive, dynamic state and determined its structure at 3.5-angstrom resolution using cryo-electron microscopy. A 180° rotation and 100-angstrom displacement of the kinase domain fully exposes the AL. On the basis of the structure and supporting biophysical data, we propose a multistep mechanism explaining how adenine nucleotides and pharmacological agonists modulate AMPK activity by altering AL phosphorylation and accessibility.
