EMD-2513
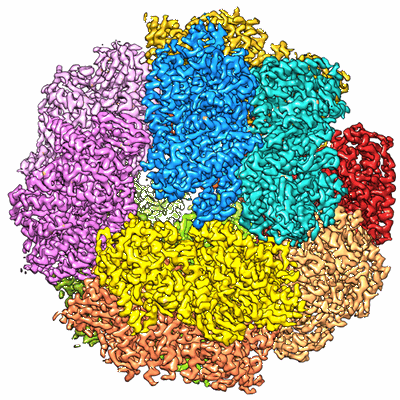
Electron cryo-microscopy of F420-reducing [NiFe] hydrogenase Frh
EMD-2513
Single-particle3.36 Å
 Deposition: 21/11/2013
Deposition: 21/11/2013Map released: 26/02/2014
Last modified: 17/02/2016
Sample Organism:
Methanothermobacter marburgensis
Sample: F420 reducing hydrogenase
Fitted models: 4ci0 (Avg. Q-score: 0.511)
Deposition Authors: Allegretti M ,
Mills DJ
,
Mills DJ  ,
McMullan G,
Kuehlbrandt W
,
McMullan G,
Kuehlbrandt W  ,
Vonck J
,
Vonck J 
Sample: F420 reducing hydrogenase
Fitted models: 4ci0 (Avg. Q-score: 0.511)
Deposition Authors: Allegretti M
 ,
Mills DJ
,
Mills DJ  ,
McMullan G,
Kuehlbrandt W
,
McMullan G,
Kuehlbrandt W  ,
Vonck J
,
Vonck J 
Atomic model of the F420-reducing [NiFe] hydrogenase by electron cryo-microscopy using a direct electron detector.
Abstract:
The introduction of direct electron detectors with higher detective quantum efficiency and fast read-out marks the beginning of a new era in electron cryo-microscopy. Using the FEI Falcon II direct electron detector in video mode, we have reconstructed a map at 3.36 Å resolution of the 1.2 MDa F420-reducing hydrogenase (Frh) from methanogenic archaea from only 320,000 asymmetric units. Videos frames were aligned by a combination of image and particle alignment procedures to overcome the effects of beam-induced motion. The reconstructed density map shows all secondary structure as well as clear side chain densities for most residues. The full coordination of all cofactors in the electron transfer chain (a [NiFe] center, four [4Fe4S] clusters and an FAD) is clearly visible along with a well-defined substrate access channel. From the rigidity of the complex we conclude that catalysis is diffusion-limited and does not depend on protein flexibility or conformational changes. DOI: http://dx.doi.org/10.7554/eLife.01963.001.
The introduction of direct electron detectors with higher detective quantum efficiency and fast read-out marks the beginning of a new era in electron cryo-microscopy. Using the FEI Falcon II direct electron detector in video mode, we have reconstructed a map at 3.36 Å resolution of the 1.2 MDa F420-reducing hydrogenase (Frh) from methanogenic archaea from only 320,000 asymmetric units. Videos frames were aligned by a combination of image and particle alignment procedures to overcome the effects of beam-induced motion. The reconstructed density map shows all secondary structure as well as clear side chain densities for most residues. The full coordination of all cofactors in the electron transfer chain (a [NiFe] center, four [4Fe4S] clusters and an FAD) is clearly visible along with a well-defined substrate access channel. From the rigidity of the complex we conclude that catalysis is diffusion-limited and does not depend on protein flexibility or conformational changes. DOI: http://dx.doi.org/10.7554/eLife.01963.001.
