EMD-25736
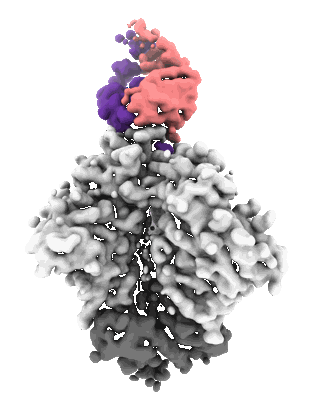
HIV-1 Envelope ApexGT3.N130 in complex with PG9 Fab
EMD-25736
Single-particle4.75 Å
 Deposition: 14/12/2021
Deposition: 14/12/2021Map released: 28/09/2022
Last modified: 06/11/2024
Sample Organism:
Human immunodeficiency virus 1,
Homo sapiens
Sample: HIV-1 Envelope ApexGT3.N130 in complex with PG9 Fab
Fitted models: 7t77 (Avg. Q-score: 0.282)
Deposition Authors: Berndsen ZT ,
Ward AB
,
Ward AB 
Sample: HIV-1 Envelope ApexGT3.N130 in complex with PG9 Fab
Fitted models: 7t77 (Avg. Q-score: 0.282)
Deposition Authors: Berndsen ZT
 ,
Ward AB
,
Ward AB 
Human immunoglobulin repertoire analysis guides design of vaccine priming immunogens targeting HIV V2-apex broadly neutralizing antibody precursors.
Willis JR,
Berndsen ZT  ,
Ma KM,
Steichen JM,
Schiffner T,
Landais E,
Liguori A,
Kalyuzhniy O,
Allen JD
,
Ma KM,
Steichen JM,
Schiffner T,
Landais E,
Liguori A,
Kalyuzhniy O,
Allen JD  ,
Baboo S
,
Baboo S  ,
Omorodion O
,
Omorodion O  ,
Diedrich JK,
Hu X,
Georgeson E,
Phelps N,
Eskandarzadeh S,
Groschel B,
Kubitz M,
Adachi Y,
Mullin TM,
Alavi NB,
Falcone S,
Himansu S,
Carfi A,
Wilson IA,
Yates 3rd JR,
Paulson JC,
Crispin M
,
Diedrich JK,
Hu X,
Georgeson E,
Phelps N,
Eskandarzadeh S,
Groschel B,
Kubitz M,
Adachi Y,
Mullin TM,
Alavi NB,
Falcone S,
Himansu S,
Carfi A,
Wilson IA,
Yates 3rd JR,
Paulson JC,
Crispin M  ,
Ward AB
,
Ward AB  ,
Schief WR
,
Schief WR
(2022) Immunity , 55 , 2149
 ,
Ma KM,
Steichen JM,
Schiffner T,
Landais E,
Liguori A,
Kalyuzhniy O,
Allen JD
,
Ma KM,
Steichen JM,
Schiffner T,
Landais E,
Liguori A,
Kalyuzhniy O,
Allen JD  ,
Baboo S
,
Baboo S  ,
Omorodion O
,
Omorodion O  ,
Diedrich JK,
Hu X,
Georgeson E,
Phelps N,
Eskandarzadeh S,
Groschel B,
Kubitz M,
Adachi Y,
Mullin TM,
Alavi NB,
Falcone S,
Himansu S,
Carfi A,
Wilson IA,
Yates 3rd JR,
Paulson JC,
Crispin M
,
Diedrich JK,
Hu X,
Georgeson E,
Phelps N,
Eskandarzadeh S,
Groschel B,
Kubitz M,
Adachi Y,
Mullin TM,
Alavi NB,
Falcone S,
Himansu S,
Carfi A,
Wilson IA,
Yates 3rd JR,
Paulson JC,
Crispin M  ,
Ward AB
,
Ward AB  ,
Schief WR
,
Schief WR
(2022) Immunity , 55 , 2149
Abstract:
Broadly neutralizing antibodies (bnAbs) to the HIV envelope (Env) V2-apex region are important leads for HIV vaccine design. Most V2-apex bnAbs engage Env with an uncommonly long heavy-chain complementarity-determining region 3 (HCDR3), suggesting that the rarity of bnAb precursors poses a challenge for vaccine priming. We created precursor sequence definitions for V2-apex HCDR3-dependent bnAbs and searched for related precursors in human antibody heavy-chain ultradeep sequencing data from 14 HIV-unexposed donors. We found potential precursors in a majority of donors for only two long-HCDR3 V2-apex bnAbs, PCT64 and PG9, identifying these bnAbs as priority vaccine targets. We then engineered ApexGT Env trimers that bound inferred germlines for PCT64 and PG9 and had higher affinities for bnAbs, determined cryo-EM structures of ApexGT trimers complexed with inferred-germline and bnAb forms of PCT64 and PG9, and developed an mRNA-encoded cell-surface ApexGT trimer. These methods and immunogens have promise to assist HIV vaccine development.
Broadly neutralizing antibodies (bnAbs) to the HIV envelope (Env) V2-apex region are important leads for HIV vaccine design. Most V2-apex bnAbs engage Env with an uncommonly long heavy-chain complementarity-determining region 3 (HCDR3), suggesting that the rarity of bnAb precursors poses a challenge for vaccine priming. We created precursor sequence definitions for V2-apex HCDR3-dependent bnAbs and searched for related precursors in human antibody heavy-chain ultradeep sequencing data from 14 HIV-unexposed donors. We found potential precursors in a majority of donors for only two long-HCDR3 V2-apex bnAbs, PCT64 and PG9, identifying these bnAbs as priority vaccine targets. We then engineered ApexGT Env trimers that bound inferred germlines for PCT64 and PG9 and had higher affinities for bnAbs, determined cryo-EM structures of ApexGT trimers complexed with inferred-germline and bnAb forms of PCT64 and PG9, and developed an mRNA-encoded cell-surface ApexGT trimer. These methods and immunogens have promise to assist HIV vaccine development.
