EMD-9103
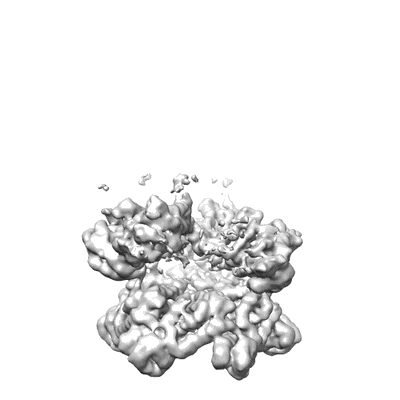
The D1 and D2 domain rings of NSF engaging the SNAP-25 N-terminus within the 20S supercomplex (focused refinement on D1/D2 rings, class 2)
EMD-9103
Single-particle3.8 Å
 Deposition: 04/09/2018
Deposition: 04/09/2018Map released: 19/09/2018
Last modified: 13/03/2024
Sample Organism:
Rattus norvegicus,
Cricetulus griseus
Sample: 20S supercomplex consisting of soluble neuronal SNARE complex, alpha-SNAP, and N-ethylmaleimide sensitive factor (NSF)
Fitted models: 6mdp (Avg. Q-score: 0.316)
Deposition Authors: White KI ,
Zhao M
,
Zhao M 
Sample: 20S supercomplex consisting of soluble neuronal SNARE complex, alpha-SNAP, and N-ethylmaleimide sensitive factor (NSF)
Fitted models: 6mdp (Avg. Q-score: 0.316)
Deposition Authors: White KI
 ,
Zhao M
,
Zhao M 
Structural principles of SNARE complex recognition by the AAA+ protein NSF.
Abstract:
The recycling of SNARE proteins following complex formation and membrane fusion is an essential process in eukaryotic trafficking. A highly conserved AAA+ protein, NSF (N-ethylmaleimide sensitive factor) and an adaptor protein, SNAP (soluble NSF attachment protein), disassemble the SNARE complex. We report electron-cryomicroscopy structures of the complex of NSF, αSNAP, and the full-length soluble neuronal SNARE complex (composed of syntaxin-1A, synaptobrevin-2, SNAP-25A) in the presence of ATP under non-hydrolyzing conditions at ~3.9 Å resolution. These structures reveal electrostatic interactions by which two αSNAP molecules interface with a specific surface of the SNARE complex. This interaction positions the SNAREs such that the 15 N-terminal residues of SNAP-25A are loaded into the D1 ring pore of NSF via a spiral pattern of interactions between a conserved tyrosine NSF residue and SNAP-25A backbone atoms. This loading process likely precedes ATP hydrolysis. Subsequent ATP hydrolysis then drives complete disassembly.
The recycling of SNARE proteins following complex formation and membrane fusion is an essential process in eukaryotic trafficking. A highly conserved AAA+ protein, NSF (N-ethylmaleimide sensitive factor) and an adaptor protein, SNAP (soluble NSF attachment protein), disassemble the SNARE complex. We report electron-cryomicroscopy structures of the complex of NSF, αSNAP, and the full-length soluble neuronal SNARE complex (composed of syntaxin-1A, synaptobrevin-2, SNAP-25A) in the presence of ATP under non-hydrolyzing conditions at ~3.9 Å resolution. These structures reveal electrostatic interactions by which two αSNAP molecules interface with a specific surface of the SNARE complex. This interaction positions the SNAREs such that the 15 N-terminal residues of SNAP-25A are loaded into the D1 ring pore of NSF via a spiral pattern of interactions between a conserved tyrosine NSF residue and SNAP-25A backbone atoms. This loading process likely precedes ATP hydrolysis. Subsequent ATP hydrolysis then drives complete disassembly.
