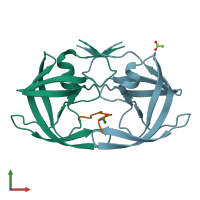
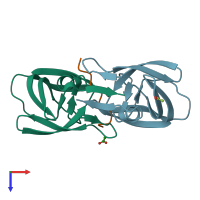
Assemblies
Assembly Name:
Protease
Multimeric state:
hetero trimer
Accessible surface area:
9020.83 Å2
Buried surface area:
6018.81 Å2
Dissociation area:
2,348.97
Å2
Dissociation energy (ΔGdiss):
34.5
kcal/mol
Dissociation entropy (TΔSdiss):
11.17
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-136844
Macromolecules
Chains: A, B
Length: 99 amino acids
Theoretical weight: 10.8 KDa
Source organism: Human immunodeficiency virus 1
Expression system: Escherichia coli
UniProt:
Pfam: Retroviral aspartyl protease
InterPro:
SCOP: Retroviral protease (retropepsin)
Length: 99 amino acids
Theoretical weight: 10.8 KDa
Source organism: Human immunodeficiency virus 1
Expression system: Escherichia coli
UniProt:
- Canonical:
 P03369 (Residues: 491-589; Coverage: 7%)
P03369 (Residues: 491-589; Coverage: 7%)
Pfam: Retroviral aspartyl protease
InterPro:
- Aspartic peptidase domain superfamily
- Retropepsins
- Retropepsin-like catalytic domain
- Peptidase A2A, retrovirus, catalytic
SCOP: Retroviral protease (retropepsin)
P03369
Chains
Domains
Secondary structure
Flexibility predictions
Early folding residue predictions
Ligand binding sites
Interaction interfaces
Chain: P
Length: 10 amino acids
Theoretical weight: 1.19 KDa
Source organism: Human immunodeficiency virus type 1 BH10
Expression system: Not provided
UniProt:
Length: 10 amino acids
Theoretical weight: 1.19 KDa
Source organism: Human immunodeficiency virus type 1 BH10
Expression system: Not provided
UniProt:
- Canonical:
 P03366 (Residues: 1155-1164; Coverage: 1%)
P03366 (Residues: 1155-1164; Coverage: 1%)
Loading Protvista...