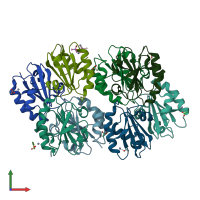
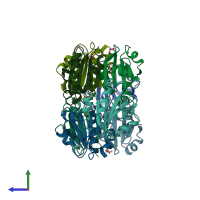
Assemblies
Assembly Name:
Thioredoxin
Multimeric state:
homo tetramer
Accessible surface area:
17846.92 Å2
Buried surface area:
6710.34 Å2
Dissociation area:
2,418.41
Å2
Dissociation energy (ΔGdiss):
7.45
kcal/mol
Dissociation entropy (TΔSdiss):
34.29
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-160431
Assembly Name:
Thioredoxin
Multimeric state:
homo tetramer
Accessible surface area:
17879.67 Å2
Buried surface area:
6286.35 Å2
Dissociation area:
2,466.49
Å2
Dissociation energy (ΔGdiss):
5.86
kcal/mol
Dissociation entropy (TΔSdiss):
34.23
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-160431
Assembly Name:
Thioredoxin
Multimeric state:
homo octamer
Accessible surface area:
33336.58 Å2
Buried surface area:
15386.7 Å2
Dissociation area:
5,031.13
Å2
Dissociation energy (ΔGdiss):
24.99
kcal/mol
Dissociation entropy (TΔSdiss):
61.02
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-160432
Assembly Name:
Thioredoxin
Multimeric state:
homo octamer
Accessible surface area:
34974.87 Å2
Buried surface area:
13748.42 Å2
Dissociation area:
3,059.76
Å2
Dissociation energy (ΔGdiss):
8.91
kcal/mol
Dissociation entropy (TΔSdiss):
36.8
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-160432
Assembly Name:
Thioredoxin
Multimeric state:
homo octamer
Accessible surface area:
35022.79 Å2
Buried surface area:
13710.34 Å2
Dissociation area:
402.72
Å2
Dissociation energy (ΔGdiss):
6.18
kcal/mol
Dissociation entropy (TΔSdiss):
12.7
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-160432
Assembly Name:
Thioredoxin
Multimeric state:
homo octamer
Accessible surface area:
35274.14 Å2
Buried surface area:
13449.14 Å2
Dissociation area:
5,125.95
Å2
Dissociation energy (ΔGdiss):
0.73
kcal/mol
Dissociation entropy (TΔSdiss):
81.34
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-160432
Macromolecules
Chains: A, B, C, D, E, F, G, H
Length: 105 amino acids
Theoretical weight: 11.56 KDa
Source organism: Alicyclobacillus acidocaldarius
Expression system: Escherichia coli
UniProt:
Pfam: Thioredoxin
InterPro:
CATH: Glutaredoxin
SCOP: Thioltransferase
Length: 105 amino acids
Theoretical weight: 11.56 KDa
Source organism: Alicyclobacillus acidocaldarius
Expression system: Escherichia coli
UniProt:
- Canonical:
 P80579 (Residues: 1-105; Coverage: 100%)
P80579 (Residues: 1-105; Coverage: 100%)
Pfam: Thioredoxin
InterPro:
CATH: Glutaredoxin
SCOP: Thioltransferase