Assemblies
Assembly Name:
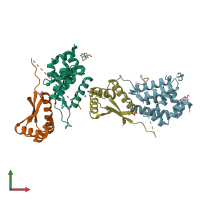
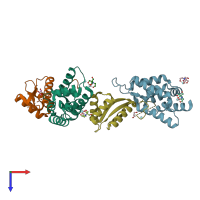
ATP-dependent Clp protease ATP-binding subunit ClpA and ATP-dependent Clp protease adapter protein ClpS
Multimeric state:
hetero dimer
Accessible surface area:
12639.02 Å2
Buried surface area:
3568.33 Å2
Dissociation area:
1,391.46
Å2
Dissociation energy (ΔGdiss):
13.81
kcal/mol
Dissociation entropy (TΔSdiss):
11.53
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-141727
Assembly Name:
ATP-dependent Clp protease ATP-binding subunit ClpA and ATP-dependent Clp protease adapter protein ClpS
Multimeric state:
hetero dimer
Accessible surface area:
12890.65 Å2
Buried surface area:
3350.53 Å2
Dissociation area:
1,274.48
Å2
Dissociation energy (ΔGdiss):
9.83
kcal/mol
Dissociation entropy (TΔSdiss):
11.34
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-141727
Assembly Name:
ATP-dependent Clp protease ATP-binding subunit ClpA and ATP-dependent Clp protease adapter protein ClpS
Multimeric state:
hetero tetramer
Accessible surface area:
24233.49 Å2
Buried surface area:
8215.05 Å2
Dissociation area:
668.78
Å2
Dissociation energy (ΔGdiss):
0.26
kcal/mol
Dissociation entropy (TΔSdiss):
12.54
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-141733
Macromolecules
Chains: A, B
Length: 143 amino acids
Theoretical weight: 16.2 KDa
Source organism: Escherichia coli
Expression system: Escherichia coli
UniProt:
Pfam: Clp amino terminal domain, pathogenicity island component
InterPro:
CATH: Clp, N-terminal domain
SCOP: Double Clp-N motif
Length: 143 amino acids
Theoretical weight: 16.2 KDa
Source organism: Escherichia coli
Expression system: Escherichia coli
UniProt:
- Canonical:
 P0ABH9 (Residues: 1-143; Coverage: 19%)
P0ABH9 (Residues: 1-143; Coverage: 19%)
Pfam: Clp amino terminal domain, pathogenicity island component
InterPro:
CATH: Clp, N-terminal domain
SCOP: Double Clp-N motif
Chains: C, D
Length: 106 amino acids
Theoretical weight: 12.19 KDa
Source organism: Escherichia coli
Expression system: Escherichia coli
UniProt:
Pfam: ATP-dependent Clp protease adaptor protein ClpS
InterPro:
SCOP: Adaptor protein ClpS (YljA)
Length: 106 amino acids
Theoretical weight: 12.19 KDa
Source organism: Escherichia coli
Expression system: Escherichia coli
UniProt:
- Canonical:
 P0A8Q6 (Residues: 1-106; Coverage: 100%)
P0A8Q6 (Residues: 1-106; Coverage: 100%)
Pfam: ATP-dependent Clp protease adaptor protein ClpS
InterPro:
- Ribosomal protein bL12, C-terminal/adaptor protein ClpS-like
- ATP-dependent Clp protease adaptor protein ClpS
- Adaptor protein ClpS, core
SCOP: Adaptor protein ClpS (YljA)