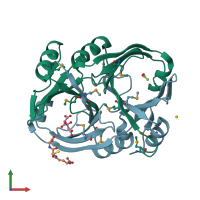
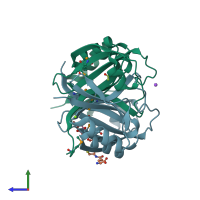
Assemblies
Assembly Name:
VOC domain-containing protein
Multimeric state:
homo dimer
Accessible surface area:
12001.26 Å2
Buried surface area:
8953.2 Å2
Dissociation area:
3,858.86
Å2
Dissociation energy (ΔGdiss):
60.03
kcal/mol
Dissociation entropy (TΔSdiss):
13.1
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-182514
Assembly Name:
VOC domain-containing protein
Multimeric state:
homo tetramer
Accessible surface area:
22796.2 Å2
Buried surface area:
18988.02 Å2
Dissociation area:
918.28
Å2
Dissociation energy (ΔGdiss):
5.37
kcal/mol
Dissociation entropy (TΔSdiss):
14.98
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-182515
Macromolecules
Chains: A, B
Length: 153 amino acids
Theoretical weight: 17.15 KDa
Source organism: Bacillus cereus
Expression system: Escherichia coli
UniProt:
Pfam: Glyoxalase/Bleomycin resistance protein/Dioxygenase superfamily
InterPro:
SCOP: Hypothetical protein BC1747
Length: 153 amino acids
Theoretical weight: 17.15 KDa
Source organism: Bacillus cereus
Expression system: Escherichia coli
UniProt:
- Canonical:
 Q81F54 (Residues: 1-150; Coverage: 100%)
Q81F54 (Residues: 1-150; Coverage: 100%)
Pfam: Glyoxalase/Bleomycin resistance protein/Dioxygenase superfamily
InterPro:
- Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenase
- Vicinal oxygen chelate (VOC) domain
SCOP: Hypothetical protein BC1747