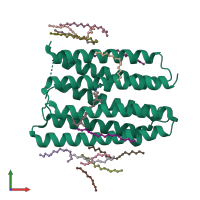Assemblies
Assembly Name:
Bacteriorhodopsin
Multimeric state:
monomeric
Accessible surface area:
10659.34 Å2
Buried surface area:
6665.04 Å2
Dissociation area:
119.06
Å2
Dissociation energy (ΔGdiss):
-3.05
kcal/mol
Dissociation entropy (TΔSdiss):
2.1
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-187105

Assembly 1
Confidence : 72%
No. subunits : 1
Symmetry : None
Assembly Name:
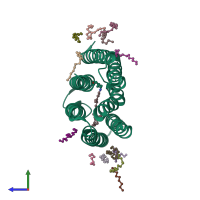
Bacteriorhodopsin
Multimeric state:
homo dimer
Accessible surface area:
17432.63 Å2
Buried surface area:
17226.23 Å2
Dissociation area:
1,846.78
Å2
Dissociation energy (ΔGdiss):
11.57
kcal/mol
Dissociation entropy (TΔSdiss):
13.3
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-187106

Assembly 2
Confidence : 89%
No. subunits : 2
Symmetry : C2
Macromolecules
Chain: A
Length: 261 amino acids
Theoretical weight: 30.22 KDa
Source organism: Nostoc sp. PCC 7120 = FACHB-418
Expression system: Escherichia coli
UniProt:
Pfam: Bacteriorhodopsin-like protein
InterPro:
CATH: Rhodopsin 7-helix transmembrane proteins
SCOP: Bacteriorhodopsin-like
Length: 261 amino acids
Theoretical weight: 30.22 KDa
Source organism: Nostoc sp. PCC 7120 = FACHB-418
Expression system: Escherichia coli
UniProt:
- Canonical:
 Q8YSC4 (Residues: 1-261; Coverage: 100%)
Q8YSC4 (Residues: 1-261; Coverage: 100%)
Pfam: Bacteriorhodopsin-like protein
InterPro:
CATH: Rhodopsin 7-helix transmembrane proteins
SCOP: Bacteriorhodopsin-like
Q8YSC4
Chains
Domains
Secondary structure
Flexibility predictions
Early folding residue predictions
Ligand binding sites