Assemblies
Multimeric state:
hetero trimer
Accessible surface area:
24928.79 Å2
Buried surface area:
11430.5 Å2
Dissociation area:
2,785.59
Å2
Dissociation energy (ΔGdiss):
29.56
kcal/mol
Dissociation entropy (TΔSdiss):
13.01
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-135943
Multimeric state:
hetero hexamer
Accessible surface area:
46785.64 Å2
Buried surface area:
25948.29 Å2
Dissociation area:
1,560.97
Å2
Dissociation energy (ΔGdiss):
6.55
kcal/mol
Dissociation entropy (TΔSdiss):
14.98
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-135945
Macromolecules
Chain: T
Length: 107 amino acids
Theoretical weight: 12.69 KDa
Source organism: Gallus gallus
Expression system: Escherichia coli BL21(DE3)
UniProt:
Pfam: Troponin
InterPro:
CATH: Single alpha-helices involved in coiled-coils or other helix-helix interfaces
SCOP: Troponin T
Length: 107 amino acids
Theoretical weight: 12.69 KDa
Source organism: Gallus gallus
Expression system: Escherichia coli BL21(DE3)
UniProt:
- Canonical:
 P12620 (Residues: 157-263; Coverage: 41%)
P12620 (Residues: 157-263; Coverage: 41%)
Pfam: Troponin
InterPro:
CATH: Single alpha-helices involved in coiled-coils or other helix-helix interfaces
SCOP: Troponin T
Chain: I
Length: 182 amino acids
Theoretical weight: 21.12 KDa
Source organism: Gallus gallus
Expression system: Escherichia coli BL21(DE3)
UniProt:
Pfam: Troponin
InterPro:
CATH: Single alpha-helices involved in coiled-coils or other helix-helix interfaces
SCOP: Troponin I
Length: 182 amino acids
Theoretical weight: 21.12 KDa
Source organism: Gallus gallus
Expression system: Escherichia coli BL21(DE3)
UniProt:
- Canonical:
 P68246 (Residues: 2-183; Coverage: 100%)
P68246 (Residues: 2-183; Coverage: 100%)
Pfam: Troponin
InterPro:
CATH: Single alpha-helices involved in coiled-coils or other helix-helix interfaces
SCOP: Troponin I
Chain: C
Length: 162 amino acids
Theoretical weight: 18.26 KDa
Source organism: Gallus gallus
Expression system: Escherichia coli BL21(DE3)
UniProt:
Pfam:
InterPro:
CATH: EF-hand
SCOP: Calmodulin-like
Length: 162 amino acids
Theoretical weight: 18.26 KDa
Source organism: Gallus gallus
Expression system: Escherichia coli BL21(DE3)
UniProt:
- Canonical:
 P02588 (Residues: 2-163; Coverage: 99%)
P02588 (Residues: 2-163; Coverage: 99%)
Pfam:
InterPro:
CATH: EF-hand
SCOP: Calmodulin-like



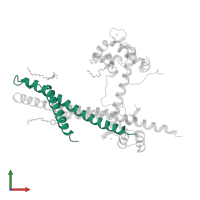
![The deposited structure of PDB entry <span class='highlight'>1ytz</span> coloured by chain, front view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>1 copy of <span class='highlight'>Troponin T, fast skeletal muscle isoforms</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>Troponin I, fast skeletal muscle</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>Troponin C, skeletal muscle</span>;</li> <li class='image_legend_li'>3 copies of <span class='highlight'>ALPHA-[4-(1,1,3,3 - TETRAMETHYLBUTYL)PHENYL]-OMEGA-HYDROXY-POLY(OXY-1,2-ETHANEDIYL)</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>CALCIUM ION</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>water</span>.</li></ul> PDB entry 1ytz coloured by chain, front view.](https://www.ebi.ac.uk/pdbe/static/entry/1ytz_deposited_chain_front_image-200x200.png)
![The deposited structure of PDB entry <span class='highlight'>1ytz</span> coloured by chain, side view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>1 copy of <span class='highlight'>Troponin T, fast skeletal muscle isoforms</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>Troponin I, fast skeletal muscle</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>Troponin C, skeletal muscle</span>;</li> <li class='image_legend_li'>3 copies of <span class='highlight'>ALPHA-[4-(1,1,3,3 - TETRAMETHYLBUTYL)PHENYL]-OMEGA-HYDROXY-POLY(OXY-1,2-ETHANEDIYL)</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>CALCIUM ION</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>water</span>.</li></ul> PDB entry 1ytz coloured by chain, side view.](https://www.ebi.ac.uk/pdbe/static/entry/1ytz_deposited_chain_side_image-200x200.png)
![The deposited structure of PDB entry <span class='highlight'>1ytz</span> coloured by chain, top view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>1 copy of <span class='highlight'>Troponin T, fast skeletal muscle isoforms</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>Troponin I, fast skeletal muscle</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>Troponin C, skeletal muscle</span>;</li> <li class='image_legend_li'>3 copies of <span class='highlight'>ALPHA-[4-(1,1,3,3 - TETRAMETHYLBUTYL)PHENYL]-OMEGA-HYDROXY-POLY(OXY-1,2-ETHANEDIYL)</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>CALCIUM ION</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>water</span>.</li></ul> PDB entry 1ytz coloured by chain, top view.](https://www.ebi.ac.uk/pdbe/static/entry/1ytz_deposited_chain_top_image-200x200.png)
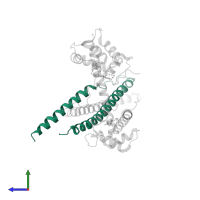
![Hetero trimeric assembly 1 of PDB entry <span class='highlight'>1ytz</span> coloured by chemically distinct molecules, front view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>1 copy of <span class='highlight'>Troponin T, fast skeletal muscle isoforms</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>Troponin I, fast skeletal muscle</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>Troponin C, skeletal muscle</span>;</li> <li class='image_legend_li'>3 copies of <span class='highlight'>ALPHA-[4-(1,1,3,3 - TETRAMETHYLBUTYL)PHENYL]-OMEGA-HYDROXY-POLY(OXY-1,2-ETHANEDIYL)</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>CALCIUM ION</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>water</span>.</li></ul>} Hetero trimeric assembly 1 of PDB entry 1ytz coloured by chemically distinct molecules, front view.](https://www.ebi.ac.uk/pdbe/static/entry/1ytz_assembly_1_chemically_distinct_molecules_front_image-200x200.png)
![Hetero trimeric assembly 1 of PDB entry <span class='highlight'>1ytz</span> coloured by chemically distinct molecules, side view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>1 copy of <span class='highlight'>Troponin T, fast skeletal muscle isoforms</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>Troponin I, fast skeletal muscle</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>Troponin C, skeletal muscle</span>;</li> <li class='image_legend_li'>3 copies of <span class='highlight'>ALPHA-[4-(1,1,3,3 - TETRAMETHYLBUTYL)PHENYL]-OMEGA-HYDROXY-POLY(OXY-1,2-ETHANEDIYL)</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>CALCIUM ION</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>water</span>.</li></ul>} Hetero trimeric assembly 1 of PDB entry 1ytz coloured by chemically distinct molecules, side view.](https://www.ebi.ac.uk/pdbe/static/entry/1ytz_assembly_1_chemically_distinct_molecules_side_image-200x200.png)
![Hetero trimeric assembly 1 of PDB entry <span class='highlight'>1ytz</span> coloured by chemically distinct molecules, top view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>1 copy of <span class='highlight'>Troponin T, fast skeletal muscle isoforms</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>Troponin I, fast skeletal muscle</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>Troponin C, skeletal muscle</span>;</li> <li class='image_legend_li'>3 copies of <span class='highlight'>ALPHA-[4-(1,1,3,3 - TETRAMETHYLBUTYL)PHENYL]-OMEGA-HYDROXY-POLY(OXY-1,2-ETHANEDIYL)</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>CALCIUM ION</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>water</span>.</li></ul>} Hetero trimeric assembly 1 of PDB entry 1ytz coloured by chemically distinct molecules, top view.](https://www.ebi.ac.uk/pdbe/static/entry/1ytz_assembly_1_chemically_distinct_molecules_top_image-200x200.png)
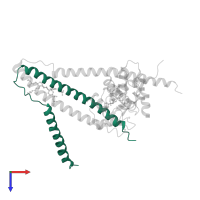
![Hetero hexameric assembly 2 of PDB entry <span class='highlight'>1ytz</span> coloured by chemically distinct molecules, front view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>2 copies of <span class='highlight'>Troponin T, fast skeletal muscle isoforms</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>Troponin I, fast skeletal muscle</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>Troponin C, skeletal muscle</span>;</li> <li class='image_legend_li'>6 copies of <span class='highlight'>ALPHA-[4-(1,1,3,3 - TETRAMETHYLBUTYL)PHENYL]-OMEGA-HYDROXY-POLY(OXY-1,2-ETHANEDIYL)</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>CALCIUM ION</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>water</span>.</li></ul>} Hetero hexameric assembly 2 of PDB entry 1ytz coloured by chemically distinct molecules, front view.](https://www.ebi.ac.uk/pdbe/static/entry/1ytz_assembly_2_chemically_distinct_molecules_front_image-200x200.png)
![Hetero hexameric assembly 2 of PDB entry <span class='highlight'>1ytz</span> coloured by chemically distinct molecules, side view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>2 copies of <span class='highlight'>Troponin T, fast skeletal muscle isoforms</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>Troponin I, fast skeletal muscle</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>Troponin C, skeletal muscle</span>;</li> <li class='image_legend_li'>6 copies of <span class='highlight'>ALPHA-[4-(1,1,3,3 - TETRAMETHYLBUTYL)PHENYL]-OMEGA-HYDROXY-POLY(OXY-1,2-ETHANEDIYL)</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>CALCIUM ION</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>water</span>.</li></ul>} Hetero hexameric assembly 2 of PDB entry 1ytz coloured by chemically distinct molecules, side view.](https://www.ebi.ac.uk/pdbe/static/entry/1ytz_assembly_2_chemically_distinct_molecules_side_image-200x200.png)
![Hetero hexameric assembly 2 of PDB entry <span class='highlight'>1ytz</span> coloured by chemically distinct molecules, top view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>2 copies of <span class='highlight'>Troponin T, fast skeletal muscle isoforms</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>Troponin I, fast skeletal muscle</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>Troponin C, skeletal muscle</span>;</li> <li class='image_legend_li'>6 copies of <span class='highlight'>ALPHA-[4-(1,1,3,3 - TETRAMETHYLBUTYL)PHENYL]-OMEGA-HYDROXY-POLY(OXY-1,2-ETHANEDIYL)</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>CALCIUM ION</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>water</span>.</li></ul>} Hetero hexameric assembly 2 of PDB entry 1ytz coloured by chemically distinct molecules, top view.](https://www.ebi.ac.uk/pdbe/static/entry/1ytz_assembly_2_chemically_distinct_molecules_top_image-200x200.png)