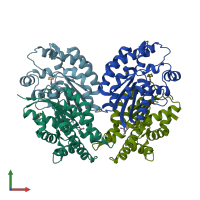
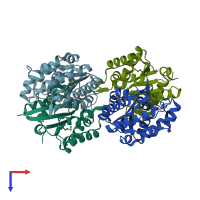
Assemblies
Assembly Name:
SGNH hydrolase-type esterase domain-containing protein
Multimeric state:
homo dimer
Accessible surface area:
16932.48 Å2
Buried surface area:
3028.68 Å2
Dissociation area:
1,019.51
Å2
Dissociation energy (ΔGdiss):
1.61
kcal/mol
Dissociation entropy (TΔSdiss):
12.66
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-187133
Assembly Name:
SGNH hydrolase-type esterase domain-containing protein
Multimeric state:
homo dimer
Accessible surface area:
16572.66 Å2
Buried surface area:
2841.09 Å2
Dissociation area:
1,047.9
Å2
Dissociation energy (ΔGdiss):
1.97
kcal/mol
Dissociation entropy (TΔSdiss):
12.65
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-187133
Macromolecules
Chains: A, B, C, D
Length: 218 amino acids
Theoretical weight: 25.23 KDa
Source organism: Nostoc sp. PCC 7120 = FACHB-418
Expression system: Escherichia coli
UniProt:
Pfam: GDSL-like Lipase/Acylhydrolase family
InterPro:
CATH: SGNH hydrolase
SCOP: Hypothetical protein alr1529
Length: 218 amino acids
Theoretical weight: 25.23 KDa
Source organism: Nostoc sp. PCC 7120 = FACHB-418
Expression system: Escherichia coli
UniProt:
- Canonical:
 Q8YWS4 (Residues: 1-206; Coverage: 100%)
Q8YWS4 (Residues: 1-206; Coverage: 100%)
Pfam: GDSL-like Lipase/Acylhydrolase family
InterPro:
CATH: SGNH hydrolase
SCOP: Hypothetical protein alr1529