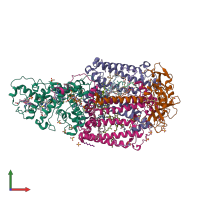
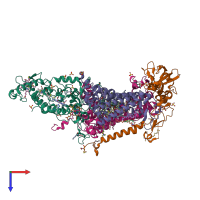
Assemblies
Multimeric state:
hetero tetramer
Accessible surface area:
39672.97 Å2
Buried surface area:
58055.9 Å2
Dissociation area:
5,837.25
Å2
Dissociation energy (ΔGdiss):
46.08
kcal/mol
Dissociation entropy (TΔSdiss):
15.44
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-138851
Macromolecules
Chain: C
Length: 336 amino acids
Theoretical weight: 37.45 KDa
Source organism: Blastochloris viridis
UniProt:
Pfam: Photosynthetic reaction centre cytochrome C subunit
InterPro:
SCOP: Photosynthetic reaction centre (cytochrome subunit)
Length: 336 amino acids
Theoretical weight: 37.45 KDa
Source organism: Blastochloris viridis
UniProt:
- Canonical:
 P07173 (Residues: 21-356; Coverage: 100%)
P07173 (Residues: 21-356; Coverage: 100%)
Pfam: Photosynthetic reaction centre cytochrome C subunit
InterPro:
- Photosynthetic reaction centre, cytochrome c subunit
- Multiheme cytochrome superfamily
- Multihaem cytochrome, PRC subunit superfamily
SCOP: Photosynthetic reaction centre (cytochrome subunit)
P07173
Chains
Domains
Secondary structure
Flexibility predictions
Early folding residue predictions
Ligand binding sites
Interaction interfaces
Sequence conservation
Chain: H
Length: 258 amino acids
Theoretical weight: 28.56 KDa
Source organism: Blastochloris viridis
UniProt:
Pfam:
InterPro:
Length: 258 amino acids
Theoretical weight: 28.56 KDa
Source organism: Blastochloris viridis
UniProt:
- Canonical:
 P06008 (Residues: 1-258; Coverage: 100%)
P06008 (Residues: 1-258; Coverage: 100%)
Pfam:
InterPro:
- Photosynthetic reaction centre, H subunit, N-terminal domain superfamily
- Photosynthetic reaction centre, H subunit
- Photosynthetic reaction centre, H subunit, N-terminal
- PRC-barrel-like superfamily
- Bacterial photosynthetic reaction centre, H-chain, C-terminal
- PRC-barrel domain
- Photosynthetic reaction centre, H subunit, N-terminal domain
- Photosynthetic Reaction Center, subunit H, domain 2
P06008
Chains
Domains
Secondary structure
Flexibility predictions
Early folding residue predictions
Ligand binding sites
Interaction interfaces
Sequence conservation
Chain: L
Length: 273 amino acids
Theoretical weight: 30.47 KDa
Source organism: Blastochloris viridis
UniProt:
Pfam: Photosynthetic reaction centre protein
InterPro:
SCOP: Bacterial photosystem II reaction centre, L and M subunits
Length: 273 amino acids
Theoretical weight: 30.47 KDa
Source organism: Blastochloris viridis
UniProt:
- Canonical:
 P06009 (Residues: 2-274; Coverage: 100%)
P06009 (Residues: 2-274; Coverage: 100%)
Pfam: Photosynthetic reaction centre protein
InterPro:
- Photosynthetic reaction centre, L subunit
- Photosystem II protein D1/D2 superfamily
- Photosynthetic reaction centre, L/M
SCOP: Bacterial photosystem II reaction centre, L and M subunits
P06009
Chains
Domains
Secondary structure
Flexibility predictions
Early folding residue predictions
Ligand binding sites
Interaction interfaces
Sequence conservation
Chain: M
Length: 323 amino acids
Theoretical weight: 35.93 KDa
Source organism: Blastochloris viridis
UniProt:
Pfam: Photosynthetic reaction centre protein
InterPro:
SCOP: Bacterial photosystem II reaction centre, L and M subunits
Length: 323 amino acids
Theoretical weight: 35.93 KDa
Source organism: Blastochloris viridis
UniProt:
- Canonical:
 P06010 (Residues: 2-324; Coverage: 100%)
P06010 (Residues: 2-324; Coverage: 100%)
Pfam: Photosynthetic reaction centre protein
InterPro:
- Photosystem II protein D1/D2 superfamily
- Photosynthetic reaction centre, L/M
- Photosynthetic reaction centre, M subunit
SCOP: Bacterial photosystem II reaction centre, L and M subunits
P06010
Chains
Domains
Secondary structure
Flexibility predictions
Early folding residue predictions
Ligand binding sites
Interaction interfaces
Sequence conservation