Assemblies
Assembly Name:
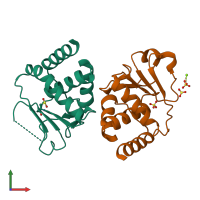
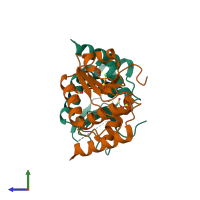
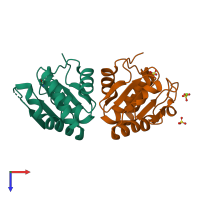
Dual-specificity protein phosphatase SDP1
Multimeric state:
monomeric
Accessible surface area:
7231.6 Å2
Buried surface area:
0.0 Å2
Dissociation area:
0
Å2
Dissociation energy (ΔGdiss):
0
kcal/mol
Dissociation entropy (TΔSdiss):
0
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-154121
Assembly Name:
Dual-specificity protein phosphatase SDP1
Multimeric state:
monomeric
Accessible surface area:
7443.79 Å2
Buried surface area:
602.48 Å2
Dissociation area:
50.76
Å2
Dissociation energy (ΔGdiss):
5.12
kcal/mol
Dissociation entropy (TΔSdiss):
0.58
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-154121
Macromolecules
Chain: A
Length: 182 amino acids
Theoretical weight: 20.9 KDa
Source organism: Saccharomyces cerevisiae
Expression system: Escherichia coli
UniProt:
Pfam: Dual specificity phosphatase, catalytic domain
InterPro:
Length: 182 amino acids
Theoretical weight: 20.9 KDa
Source organism: Saccharomyces cerevisiae
Expression system: Escherichia coli
UniProt:
- Canonical:
 P40479 (Residues: 17-198; Coverage: 87%)
P40479 (Residues: 17-198; Coverage: 87%)
Pfam: Dual specificity phosphatase, catalytic domain
InterPro:
- Protein-tyrosine phosphatase-like
- Dual specificity protein phosphatase domain
- Dual specificity phosphatase, catalytic domain
- Tyrosine-specific protein phosphatases domain
- Protein-tyrosine phosphatase, active site
P40479
Chains
Domains
Secondary structure
Flexibility predictions
Early folding residue predictions
Chain: B
Length: 182 amino acids
Theoretical weight: 20.87 KDa
Source organism: Saccharomyces cerevisiae
Expression system: Escherichia coli
UniProt:
Pfam: Dual specificity phosphatase, catalytic domain
InterPro:
Length: 182 amino acids
Theoretical weight: 20.87 KDa
Source organism: Saccharomyces cerevisiae
Expression system: Escherichia coli
UniProt:
- Canonical:
 P40479 (Residues: 17-198; Coverage: 87%)
P40479 (Residues: 17-198; Coverage: 87%)
Pfam: Dual specificity phosphatase, catalytic domain
InterPro:
- Protein-tyrosine phosphatase-like
- Dual specificity protein phosphatase domain
- Dual specificity phosphatase, catalytic domain
- Tyrosine-specific protein phosphatases domain
- Protein-tyrosine phosphatase, active site
Loading Protvista...