Assemblies
Assembly Name:
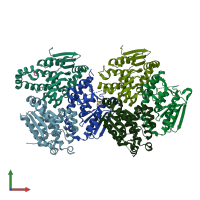
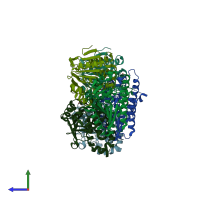
Enoyl-CoA hydratase
Multimeric state:
homo trimer
Accessible surface area:
26843.08 Å2
Buried surface area:
6916.17 Å2
Dissociation area:
3,135.27
Å2
Dissociation energy (ΔGdiss):
34.98
kcal/mol
Dissociation entropy (TΔSdiss):
26.51
kcal/mol
Symmetry number:
3
PDBe Complex ID:
PDB-CPX-495654
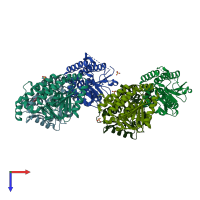
Assembly Name:
Enoyl-CoA hydratase
Multimeric state:
homo trimer
Accessible surface area:
24953.71 Å2
Buried surface area:
6532.04 Å2
Dissociation area:
3,102.09
Å2
Dissociation energy (ΔGdiss):
32.6
kcal/mol
Dissociation entropy (TΔSdiss):
26.25
kcal/mol
Symmetry number:
3
PDBe Complex ID:
PDB-CPX-495654
Macromolecules
Chains: A, B, C, D, E, F
Length: 266 amino acids
Theoretical weight: 28.99 KDa
Source organism: Acinetobacter baumannii ATCC 17978
Expression system: Escherichia coli
UniProt:
Pfam: Enoyl-CoA hydratase/isomerase
InterPro:
Length: 266 amino acids
Theoretical weight: 28.99 KDa
Source organism: Acinetobacter baumannii ATCC 17978
Expression system: Escherichia coli
UniProt:
- Canonical:
 A0A1E3M6I3 (Residues: 10-264; Coverage: 96%)
A0A1E3M6I3 (Residues: 10-264; Coverage: 96%)
Pfam: Enoyl-CoA hydratase/isomerase
InterPro:
- ClpP/crotonase-like domain superfamily
- Enoyl-CoA hydratase/isomerase
- Enoyl-CoA hydratase, C-terminal