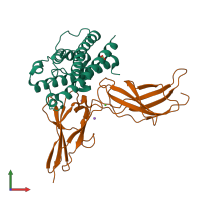
Assemblies
Assembly Name:
Prolactin receptor and Prolactin
Multimeric state:
hetero dimer
Accessible surface area:
20049.77 Å2
Buried surface area:
3061.27 Å2
Dissociation area:
1,196.68
Å2
Dissociation energy (ΔGdiss):
7.61
kcal/mol
Dissociation entropy (TΔSdiss):
12.59
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-134124
Macromolecules
Chain: A
Length: 186 amino acids
Theoretical weight: 21.72 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli BL21(DE3)
UniProt:
Pfam: Somatotropin hormone family
InterPro:
CATH: Growth Hormone; Chain: A;
Length: 186 amino acids
Theoretical weight: 21.72 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli BL21(DE3)
UniProt:
- Canonical:
 P01236 (Residues: 43-227; Coverage: 93%)
P01236 (Residues: 43-227; Coverage: 93%)
Pfam: Somatotropin hormone family
InterPro:
CATH: Growth Hormone; Chain: A;
P01236
Chains
Domains
Secondary structure
Flexibility predictions
Early folding residue predictions
Ligand binding sites
Interaction interfaces
Sequence conservation
Variants
Chain: B
Length: 210 amino acids
Theoretical weight: 24.45 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli BL21(DE3)
UniProt:
Pfam: Erythropoietin receptor, ligand binding
InterPro:
Length: 210 amino acids
Theoretical weight: 24.45 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli BL21(DE3)
UniProt:
- Canonical:
 P16471 (Residues: 25-234; Coverage: 35%)
P16471 (Residues: 25-234; Coverage: 35%)
Pfam: Erythropoietin receptor, ligand binding
InterPro:
- Immunoglobulin-like fold
- Growth hormone/erythropoietin receptor, ligand binding
- Fibronectin type III
- Fibronectin type III superfamily
- Long hematopoietin receptor, single chain, conserved site
P16471
Chains
Domains
Secondary structure
Flexibility predictions
Early folding residue predictions
Ligand binding sites
Interaction interfaces
Sequence conservation
Variants