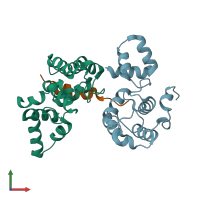Assemblies
Assembly Name:
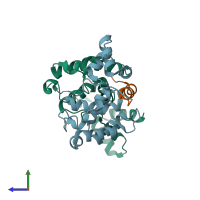
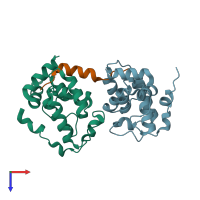
DNA-binding protein RAP1 and Regulatory protein SIR3
Multimeric state:
hetero trimer
Accessible surface area:
16879.68 Å2
Buried surface area:
2720.93 Å2
Dissociation area:
519.29
Å2
Dissociation energy (ΔGdiss):
-3
kcal/mol
Dissociation entropy (TΔSdiss):
11.71
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-139139
Macromolecules
Chains: A, B
Length: 157 amino acids
Theoretical weight: 18.22 KDa
Source organism: Saccharomyces cerevisiae
Expression system: Escherichia coli
UniProt:
Pfam: TRF2-interacting telomeric protein/Rap1 - C terminal domain
InterPro:
Length: 157 amino acids
Theoretical weight: 18.22 KDa
Source organism: Saccharomyces cerevisiae
Expression system: Escherichia coli
UniProt:
- Canonical:
 P11938 (Residues: 672-827; Coverage: 19%)
P11938 (Residues: 672-827; Coverage: 19%)
Pfam: TRF2-interacting telomeric protein/Rap1 - C terminal domain
InterPro:
- TE2IP/Rap1
- TRF2-interacting telomeric protein/Rap1, C-terminal
- TRF2-interacting telomeric protein/Rap1, C-terminal domain superfamily