Assemblies
Assembly Name:
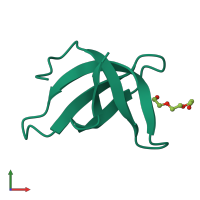
Tudor domain-containing protein 3
Multimeric state:
monomeric
Accessible surface area:
3698.02 Å2
Buried surface area:
292.96 Å2
Dissociation area:
146.48
Å2
Dissociation energy (ΔGdiss):
-4.08
kcal/mol
Dissociation entropy (TΔSdiss):
2.33
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-190662
Macromolecules
Chain: A
Length: 59 amino acids
Theoretical weight: 6.88 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli BL21(DE3)
UniProt:
Pfam: Tudor domain
InterPro:
CATH: SH3 type barrels.
Length: 59 amino acids
Theoretical weight: 6.88 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli BL21(DE3)
UniProt:
- Canonical:
 Q9H7E2 (Residues: 553-611; Coverage: 9%)
Q9H7E2 (Residues: 553-611; Coverage: 9%)
Pfam: Tudor domain
InterPro:
CATH: SH3 type barrels.
Q9H7E2
Chains
Domains
Secondary structure
Flexibility predictions
Early folding residue predictions
Ligand binding sites