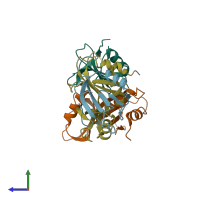
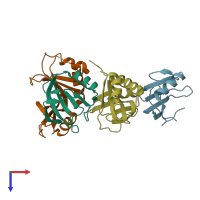
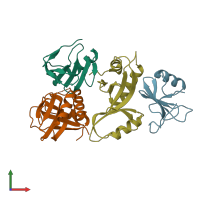Assemblies
Assembly Name:
TRCF-UvrA complex
Multimeric state:
hetero dimer
Accessible surface area:
10578.74 Å2
Buried surface area:
1749.9 Å2
Dissociation area:
874.95
Å2
Dissociation energy (ΔGdiss):
-2.76
kcal/mol
Dissociation entropy (TΔSdiss):
10.73
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-141272
Assembly Name:
TRCF-UvrA complex
Multimeric state:
hetero dimer
Accessible surface area:
10655.36 Å2
Buried surface area:
1707.73 Å2
Dissociation area:
853.86
Å2
Dissociation energy (ΔGdiss):
-2.4
kcal/mol
Dissociation entropy (TΔSdiss):
10.64
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-141272
Macromolecules
Chains: A, C
Length: 93 amino acids
Theoretical weight: 10.58 KDa
Source organism: Escherichia coli K-12
Expression system: Escherichia coli
UniProt:
Pfam: UvrB interaction domain
InterPro:
Length: 93 amino acids
Theoretical weight: 10.58 KDa
Source organism: Escherichia coli K-12
Expression system: Escherichia coli
UniProt:
- Canonical:
 P30958 (Residues: 127-213; Coverage: 8%)
P30958 (Residues: 127-213; Coverage: 8%)
Pfam: UvrB interaction domain
InterPro:
- UvrABC system, subunit B
- P-loop containing nucleoside triphosphate hydrolase
- UvrB, interaction domain
Chains: B, D
Length: 126 amino acids
Theoretical weight: 13.96 KDa
Source organism: Escherichia coli K-12
Expression system: Escherichia coli
UniProt:
Pfam: UvrA interaction domain
InterPro: UvrA, interaction domain
CATH: Ribulose 1,5 Bisphosphate Carboxylase/Oxygenase
Length: 126 amino acids
Theoretical weight: 13.96 KDa
Source organism: Escherichia coli K-12
Expression system: Escherichia coli
UniProt:
- Canonical:
 P0A698 (Residues: 131-250; Coverage: 13%)
P0A698 (Residues: 131-250; Coverage: 13%)
Pfam: UvrA interaction domain
InterPro: UvrA, interaction domain
CATH: Ribulose 1,5 Bisphosphate Carboxylase/Oxygenase