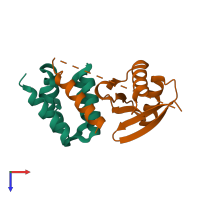Assemblies
Assembly Name:
Mitogen-activated protein kinase kinase kinase 3 and Cerebral cavernous malformations 2 protein
Multimeric state:
hetero dimer
Accessible surface area:
10143.32 Å2
Buried surface area:
2594.94 Å2
Dissociation area:
1,297.47
Å2
Dissociation energy (ΔGdiss):
10.94
kcal/mol
Dissociation entropy (TΔSdiss):
10.78
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-189267
Macromolecules
Chain: A
Length: 98 amino acids
Theoretical weight: 11.16 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli BL21(DE3)
UniProt:
Pfam: Cerebral cavernous malformation protein, harmonin-homology
InterPro:
CATH: Paired amphipathic helix 2 (pah2 repeat)
Length: 98 amino acids
Theoretical weight: 11.16 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli BL21(DE3)
UniProt:
- Canonical:
 Q9BSQ5 (Residues: 283-379; Coverage: 22%)
Q9BSQ5 (Residues: 283-379; Coverage: 22%)
Pfam: Cerebral cavernous malformation protein, harmonin-homology
InterPro:
CATH: Paired amphipathic helix 2 (pah2 repeat)
Q9BSQ5
Chains
Domains
Secondary structure
Flexibility predictions
Early folding residue predictions
Interaction interfaces
Sequence conservation
Chain: B
Length: 126 amino acids
Theoretical weight: 14.68 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli BL21(DE3)
UniProt:
Pfam: PB1 domain
InterPro:
Length: 126 amino acids
Theoretical weight: 14.68 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli BL21(DE3)
UniProt:
- Canonical:
 Q99759 (Residues: 1-124; Coverage: 20%)
Q99759 (Residues: 1-124; Coverage: 20%)
Pfam: PB1 domain
InterPro:
Q99759
Chains
Domains
Secondary structure
Flexibility predictions
Early folding residue predictions
Interaction interfaces