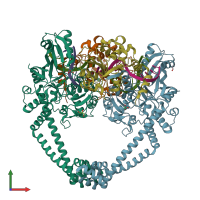
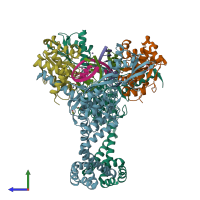
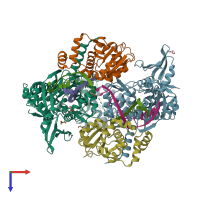
Assemblies
Assembly Name:
DNA gyrase and DNA
Multimeric state:
hetero heptamer
Accessible surface area:
57472.58 Å2
Buried surface area:
21891.67 Å2
Dissociation area:
1,310.28
Å2
Dissociation energy (ΔGdiss):
17.38
kcal/mol
Dissociation entropy (TΔSdiss):
9.4
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-118477
Macromolecules
Chains: A, C
Length: 483 amino acids
Theoretical weight: 54.74 KDa
Source organism: Staphylococcus aureus subsp. aureus N315
Expression system: Escherichia coli
UniProt:
Pfam: DNA gyrase/topoisomerase IV, subunit A
InterPro:
Length: 483 amino acids
Theoretical weight: 54.74 KDa
Source organism: Staphylococcus aureus subsp. aureus N315
Expression system: Escherichia coli
UniProt:
- Canonical:
 Q99XG5 (Residues: 9-491; Coverage: 54%)
Q99XG5 (Residues: 9-491; Coverage: 54%)
Pfam: DNA gyrase/topoisomerase IV, subunit A
InterPro:
Chains: B, D
Length: 190 amino acids
Theoretical weight: 21.35 KDa
Source organism: Staphylococcus aureus subsp. aureus N315
Expression system: Escherichia coli
UniProt:
Pfam:
InterPro:
Length: 190 amino acids
Theoretical weight: 21.35 KDa
Source organism: Staphylococcus aureus subsp. aureus N315
Expression system: Escherichia coli
UniProt:
- Canonical:
 P66937 (Residues: 417-543, 580-640; Coverage: 29%)
P66937 (Residues: 417-543, 580-640; Coverage: 29%)
Pfam:
InterPro:
Name:
DNA (5'-D(*AP*GP*CP*CP*GP*TP*AP*)-3')
Representative chains: E
Length: 8 nucleotides
Theoretical weight: 2.45 KDa
Representative chains: E
Length: 8 nucleotides
Theoretical weight: 2.45 KDa
Name:
DNA (5'-D(*GP*GP*TP*AP*CP*CP*TP*AP*CP*GP*GP*CP*T)-3')
Representative chains: G
Length: 13 nucleotides
Theoretical weight: 3.97 KDa
Representative chains: G
Length: 13 nucleotides
Theoretical weight: 3.97 KDa
Name:
DNA (5'-D(*AP*GP*CP*CP*GP*TP*AP*GP*GP*TP*AP*CP*CP*TP*AP*CP*GP*GP*CP*T)-3')
Representative chains: F
Length: 20 nucleotides
Theoretical weight: 6.13 KDa
Representative chains: F
Length: 20 nucleotides
Theoretical weight: 6.13 KDa