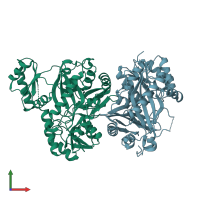
Assemblies
Assembly Name:
Biotin-dependent acyl-coenzyme A carboxylase alpha3 subunit
Multimeric state:
homo dimer
Accessible surface area:
31415.56 Å2
Buried surface area:
2424.39 Å2
Dissociation area:
1,212.19
Å2
Dissociation energy (ΔGdiss):
-11.16
kcal/mol
Dissociation entropy (TΔSdiss):
14.18
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-161259
Macromolecules
Chains: A, B
Length: 614 amino acids
Theoretical weight: 65.46 KDa
Source organism: Mycobacterium tuberculosis H37Rv
Expression system: Escherichia coli
UniProt:
Pfam:
Length: 614 amino acids
Theoretical weight: 65.46 KDa
Source organism: Mycobacterium tuberculosis H37Rv
Expression system: Escherichia coli
UniProt:
- Canonical:
 P96890 (Residues: 1-600; Coverage: 100%)
P96890 (Residues: 1-600; Coverage: 100%)
Pfam:
- Biotin carboxylase, N-terminal domain
- Carbamoyl-phosphate synthase L chain, ATP binding domain
- Biotin carboxylase C-terminal domain
- Biotin-requiring enzyme
- Pre-ATP-grasp domain superfamily
- Biotin carboxylation domain
- Biotin carboxylase-like, N-terminal domain
- Carbamoyl-phosphate synthetase large subunit-like, ATP-binding domain
- ATP-grasp fold
- ATP-grasp fold, subdomain 1
- Rudiment single hybrid motif
- Biotin carboxylase, C-terminal
- Biotin/lipoyl attachment
- Single hybrid motif
- Biotin-binding site