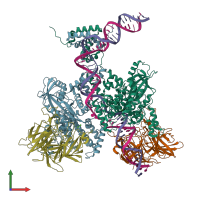
Assemblies
Multimeric state:
hetero hexamer
Accessible surface area:
98714.09 Å2
Buried surface area:
25965.53 Å2
Dissociation area:
1,801.86
Å2
Dissociation energy (ΔGdiss):
4.91
kcal/mol
Dissociation entropy (TΔSdiss):
14.9
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-119729
Macromolecules
Chains: A, C
Length: 1159 amino acids
Theoretical weight: 131.16 KDa
Source organisms: Expression system: Spodoptera frugiperda
UniProt:
Pfam:
Length: 1159 amino acids
Theoretical weight: 131.16 KDa
Source organisms: Expression system: Spodoptera frugiperda
UniProt:
- Canonical:
 P0AEX9 (Residues: 15-15, 29-392; Coverage: 95%)
P0AEX9 (Residues: 15-15, 29-392; Coverage: 95%) - Canonical:
 O13033 (Residues: 271-1031; Coverage: 72%)
nullnull
O13033 (Residues: 271-1031; Coverage: 72%)
nullnull
Pfam:
- Bacterial extracellular solute-binding protein
- Zinc finger, C3HC4 type (RING finger)
- Recombination-activation protein 1 (RAG1), recombinase
- Maltose/Cyclodextrin ABC transporter, substrate-binding protein
- Bacterial-type extracellular solute-binding protein
- Solute-binding family 1, conserved site
- V(D)J recombination-activating protein 1
- Zinc finger, RING/FYVE/PHD-type
- Zinc finger, RING-type
- Zinc finger, C3HC4 RING-type
- Zinc finger, RING-type, conserved site
- V(D)J recombination-activating protein 1, Zinc finger
- RAG nonamer-binding domain
Chains: B, D
Length: 533 amino acids
Theoretical weight: 59.44 KDa
Source organism: Danio rerio
Expression system: Spodoptera frugiperda
UniProt:
Pfam:
InterPro:
Length: 533 amino acids
Theoretical weight: 59.44 KDa
Source organism: Danio rerio
Expression system: Spodoptera frugiperda
UniProt:
- Canonical:
 O13034 (Residues: 1-530; Coverage: 100%)
O13034 (Residues: 1-530; Coverage: 100%)
Pfam:
InterPro:
Name:
Forward strand of 12-RSS substrate DNA
Representative chains: E
Length: 50 nucleotides
Theoretical weight: 15.36 KDa
Representative chains: E
Length: 50 nucleotides
Theoretical weight: 15.36 KDa
Name:
Reverse strand of 12-RSS substrate DNA
Representative chains: F
Length: 50 nucleotides
Theoretical weight: 15.44 KDa
Representative chains: F
Length: 50 nucleotides
Theoretical weight: 15.44 KDa