$VAR1 = undef;
Summary for peptidase A25.001: gpr peptidase
| Names | |
|---|---|
| MEROPS Name | gpr peptidase |
| Other names | germination protease (bacterial), gpr g.p. (Bacillus megaterium), gpr protease, spore protease |
| Domain architecture |
|---|

| MEROPS Classification | |
|---|---|
| Classification | Clan AE >> Subclan (none) >> Family A25 >> Subfamily (none) >> A25.001 |
| Holotype | gpr peptidase (Bacillus megaterium), Uniprot accession P22321 (peptidase unit: 17-371), MERNUM MER0001292 |
| History | Identifier created: MEROPS 5.61 (29 October 2001) |
| Activity | |||
|---|---|---|---|
| Catalytic type | Aspartic | ||
| Peplist | Included in the Peplist with identifier PL00263 | ||
| NC-IUBMB | Subclass 3.4 (Peptidases) >> Sub-subclass 3.4.24 (Metalloendopeptidases) >> Peptidase 3.4.24.78 | ||
| Enzymology | BRENDA database | ||
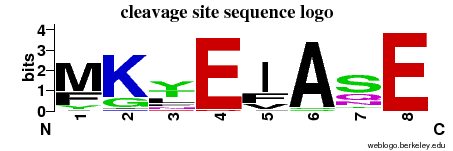
| Cleavage site specificity | Explanations of how to interpret the following cleavage site sequence logo and specificity matrix can be found here. | ||
| Cleavage pattern | Mfy/Kg/ytl/E Ifv/A/Sqn/E (based on 32 cleavages) Ifv/A/Sqn/E (based on 32 cleavages) | ||
 |
| Specificity matrix | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
