| Activity |
|---|
| Catalytic type | Serine |
| Peplist | Included in the Peplist with identifier PL00321 |
| NC-IUBMB | Subclass 3.4 (Peptidases) >> Sub-subclass 3.4.21 (Serine endopeptidases) >> Peptidase 3.4.21.7
|
| Enzymology | BRENDA database |
| Proteolytic events | CutDB database (71 cleavages) |
| Activity status | human: active (Castellino, 2004)
mouse: active (by similarity)
|
| Physiology | Degrades fibrin clots, but many additional physiologic roles have been proposed. In mouse, plasmin has been identified as an extracellular chemokine activator, enhancing the activity of monocyte chemoattractant protein-1 (MCP-1) by removal of its C-terminus (Sheehan et al., 2007). |
| Knockout | Plasminogen knockout mice showed severe thrombosis, and deposition of fibrin in the liver (Bugge et al., 1995). They also showed impaired wound-healing in the skin (Romer et al., 1996) and complete lack of infarct healing after myocardial infarction (Creemers et al., 2000). The plasminogen-deficient mice are resistant to excitotoxic neurodegeneration (Sheehan et al., 2007). |
| Pharmaceutical relevance | Microplasmin has been described as a novel thrombolytic agent that improves behavioural outcome after embolic strokes in rabbits (Lapchak %et al, 2002). An engineered, thrombin-activatable form of plasminogen has also shown promise as a thrombolytic agent (Comer et al., 2005). |
| Pathways |
KEGG | Complement and coagulation cascades |
|
KEGG | Influenza A |
|
KEGG | Neuroactive ligand-receptor interaction |
|
KEGG | Staphylococcus aureus infection |
|
Other databases
| WIKIPEDIA | http://en.wikipedia.org/wiki/Plasmin |
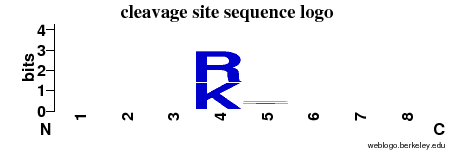
| Cleavage site specificity |
Explanations of how to interpret the
following cleavage site sequence logo and specificity matrix can be found here. |
|---|
| Cleavage pattern | -/-/-/RK -/-/-/- (based on 132 cleavages) -/-/-/- (based on 132 cleavages) |

 -/-/-/- (based on 132 cleavages)
-/-/-/- (based on 132 cleavages)