Assemblies
Assembly Name:
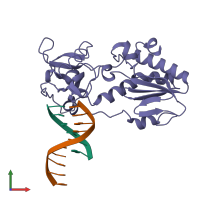
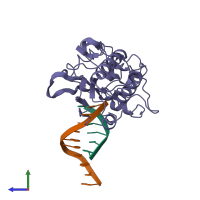
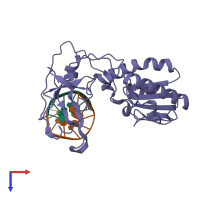
Reverse transcriptase/ribonuclease H and DNA
Multimeric state:
hetero trimer
Accessible surface area:
15565.86 Å2
Buried surface area:
1398.81 Å2
Dissociation area:
402.73
Å2
Dissociation energy (ΔGdiss):
-9.15
kcal/mol
Dissociation entropy (TΔSdiss):
10.18
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-113580
Macromolecules
Chain: A
Length: 255 amino acids
Theoretical weight: 28.93 KDa
Source organism: Moloney murine leukemia virus
Expression system: Escherichia coli
UniProt:
Pfam: Reverse transcriptase (RNA-dependent DNA polymerase)
InterPro:
SCOP: Reverse transcriptase
Length: 255 amino acids
Theoretical weight: 28.93 KDa
Source organism: Moloney murine leukemia virus
Expression system: Escherichia coli
UniProt:
- Canonical:
 P03355 (Residues: 683-937; Coverage: 15%)
P03355 (Residues: 683-937; Coverage: 15%)
Pfam: Reverse transcriptase (RNA-dependent DNA polymerase)
InterPro:
- DNA/RNA polymerase superfamily
- Reverse transcriptase/Diguanylate cyclase domain
- Reverse transcriptase domain
SCOP: Reverse transcriptase
Loading Protvista...
Name:
DNA (5'-D(*CP*TP*CP*GP*TP*G)-3')
Representative chains: B
Source organism: Moloney murine leukemia virus [11801]
Expression system: Not provided
Length: 6 nucleotides
Theoretical weight: 1.8 KDa
Representative chains: B
Source organism: Moloney murine leukemia virus [11801]
Expression system: Not provided
Length: 6 nucleotides
Theoretical weight: 1.8 KDa
Loading Protvista...
Name:
DNA (5'-D(*AP*CP*GP*GP*CP*AP*CP*GP*AP*G)-3')
Representative chains: C
Source organism: Moloney murine leukemia virus [11801]
Expression system: Not provided
Length: 10 nucleotides
Theoretical weight: 3.08 KDa
Representative chains: C
Source organism: Moloney murine leukemia virus [11801]
Expression system: Not provided
Length: 10 nucleotides
Theoretical weight: 3.08 KDa
Loading Protvista...