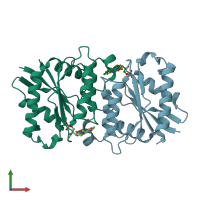
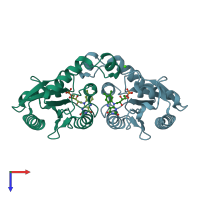
Assemblies
Assembly Name:
Quinone reductase
Multimeric state:
homo tetramer
Accessible surface area:
26056.54 Å2
Buried surface area:
12538.81 Å2
Dissociation area:
1,603.69
Å2
Dissociation energy (ΔGdiss):
8.44
kcal/mol
Dissociation entropy (TΔSdiss):
13.82
kcal/mol
Symmetry number:
4
PDBe Complex ID:
PDB-CPX-142829
Assembly Name:
Quinone reductase
Multimeric state:
homo dimer
Accessible surface area:
14617.98 Å2
Buried surface area:
4682.2 Å2
Dissociation area:
1,603.5
Å2
Dissociation energy (ΔGdiss):
11.88
kcal/mol
Dissociation entropy (TΔSdiss):
12.56
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-142828
Macromolecules
Chains: A, B
Length: 193 amino acids
Theoretical weight: 20.85 KDa
Source organism: Escherichia coli K-12
Expression system: Escherichia coli
UniProt:
Pfam: NADPH-dependent FMN reductase
InterPro:
CATH: Rossmann fold
Length: 193 amino acids
Theoretical weight: 20.85 KDa
Source organism: Escherichia coli K-12
Expression system: Escherichia coli
UniProt:
- Canonical:
 P0AGE6 (Residues: 1-188; Coverage: 100%)
P0AGE6 (Residues: 1-188; Coverage: 100%)
Pfam: NADPH-dependent FMN reductase
InterPro:
CATH: Rossmann fold