Assemblies
Assembly Name:
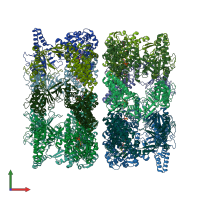
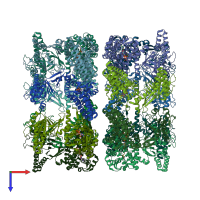
Chaperonin GroEL
Multimeric state:
homo tetradecamer
Accessible surface area:
291923.12 Å2
Buried surface area:
39717.84 Å2
Dissociation area:
4,598.17
Å2
Dissociation energy (ΔGdiss):
-43.81
kcal/mol
Dissociation entropy (TΔSdiss):
110.33
kcal/mol
Symmetry number:
7
PDBe Complex ID:
PDB-CPX-141315
Macromolecules
Chains: A, B, C, D, E, F, G, H, I, J, K, L, M, N
Length: 548 amino acids
Theoretical weight: 57.35 KDa
Source organism: Escherichia coli
Expression system: Escherichia coli
UniProt:
Pfam: TCP-1/cpn60 chaperonin family
InterPro:
Length: 548 amino acids
Theoretical weight: 57.35 KDa
Source organism: Escherichia coli
Expression system: Escherichia coli
UniProt:
- Canonical:
 P0A6F5 (Residues: 1-548; Coverage: 100%)
P0A6F5 (Residues: 1-548; Coverage: 100%)
Pfam: TCP-1/cpn60 chaperonin family
InterPro: