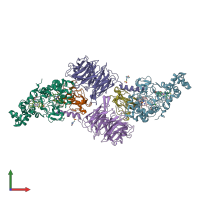
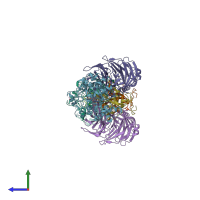
Assemblies
Multimeric state:
hetero hexamer
Accessible surface area:
58266.65 Å2
Buried surface area:
25698.47 Å2
Dissociation area:
3,086.21
Å2
Dissociation energy (ΔGdiss):
1.83
kcal/mol
Dissociation entropy (TΔSdiss):
29.04
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-106574
Macromolecules
Chains: A, B
Length: 373 amino acids
Theoretical weight: 41.13 KDa
Source organism: Paracoccus denitrificans PD1222
Expression system: Paracoccus denitrificans
UniProt:
Pfam: Di-haem cytochrome c peroxidase
InterPro:
CATH: Cytochrome c-like domain
Length: 373 amino acids
Theoretical weight: 41.13 KDa
Source organism: Paracoccus denitrificans PD1222
Expression system: Paracoccus denitrificans
UniProt:
- Canonical:
 Q51658 (Residues: 21-387; Coverage: 100%)
Q51658 (Residues: 21-387; Coverage: 100%)
Pfam: Di-haem cytochrome c peroxidase
InterPro:
CATH: Cytochrome c-like domain
Chains: C, E
Length: 137 amino acids
Theoretical weight: 15.03 KDa
Source organism: Paracoccus denitrificans PD1222
Expression system: Cereibacter sphaeroides
UniProt:
Pfam: Methylamine dehydrogenase, L chain
InterPro:
Length: 137 amino acids
Theoretical weight: 15.03 KDa
Source organism: Paracoccus denitrificans PD1222
Expression system: Cereibacter sphaeroides
UniProt:
- Canonical:
 A1BBA0 (Residues: 58-188; Coverage: 70%)
A1BBA0 (Residues: 58-188; Coverage: 70%)
Pfam: Methylamine dehydrogenase, L chain
InterPro:
- Methylamine/Aralkylamine dehydrogenase light chain superfamily
- Amine dehydrogenase light chain
- Methylamine dehydrogenase light chain
- Methylamine/Aralkylamine dehydrogenase light chain, C-terminal domain
Chains: D, F
Length: 385 amino acids
Theoretical weight: 42.32 KDa
Source organism: Paracoccus denitrificans PD1222
Expression system: Cereibacter sphaeroides
UniProt:
Pfam: Methylamine dehydrogenase heavy chain (MADH)
InterPro:
Length: 385 amino acids
Theoretical weight: 42.32 KDa
Source organism: Paracoccus denitrificans PD1222
Expression system: Cereibacter sphaeroides
UniProt:
- Canonical:
 A1BB97 (Residues: 33-417; Coverage: 99%)
A1BB97 (Residues: 33-417; Coverage: 99%)
Pfam: Methylamine dehydrogenase heavy chain (MADH)
InterPro:
- WD40/YVTN repeat-like-containing domain superfamily
- Quinoprotein amine dehydrogenase, beta chain-like
- Methylamine dehydrogenase heavy chain
- Amine dehydrogenase heavy chain